3T6G
 
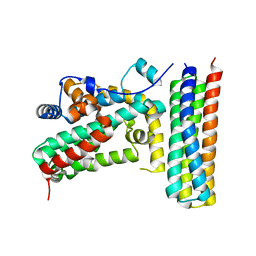 | |
3T6A
 
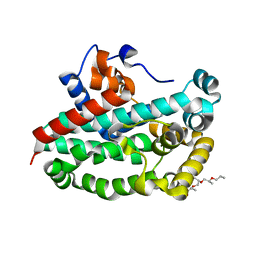 | | Structure of the C-terminal domain of BCAR3 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, Breast cancer anti-estrogen resistance protein 3, UNKNOWN ATOM OR ION | | Authors: | Mace, P.D, Robinson, H, Riedl, S.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NSP-Cas protein structures reveal a promiscuous interaction module in cell signaling.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2HLQ
 
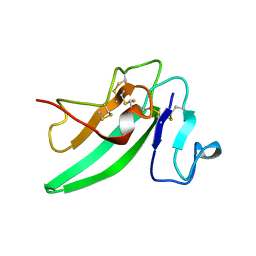 | |
2HLR
 
 | |
3M1D
 
 | | Structure of BIR1 from cIAP1 | | Descriptor: | Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Mace, P.D, Day, C.L. | | Deposit date: | 2010-03-04 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric recruitment of cIAPs by TRAF2
J.Mol.Biol., 400, 2010
|
|
4IZA
 
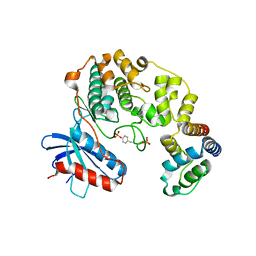 | |
4IZ5
 
 | | Structure of the complex between ERK2 phosphomimetic mutant and PEA-15 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Astrocytic phosphoprotein PEA-15, Mitogen-activated protein kinase 1, ... | | Authors: | Mace, P.D, Robinson, H, Riedl, S.J. | | Deposit date: | 2013-01-29 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of ERK2 bound to PEA-15 reveals a mechanism for rapid release of activated MAPK.
Nat Commun, 4, 2013
|
|
4IZ7
 
 | |
3EB6
 
 | | Structure of the cIAP2 RING domain bound to UbcH5b | | Descriptor: | Baculoviral IAP repeat-containing protein 3, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Mace, P.D, Linke, K, Schumacher, F.-R, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of the cIAP2 RING Domain Reveal Conformational Changes Associated with Ubiquitin-conjugating Enzyme (E2) Recruitment.
J.Biol.Chem., 283, 2008
|
|
3EB5
 
 | | Structure of the cIAP2 RING domain | | Descriptor: | Baculoviral IAP repeat-containing protein 3, SODIUM ION, ZINC ION | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment.
J.Biol.Chem., 283, 2008
|
|
2VJF
 
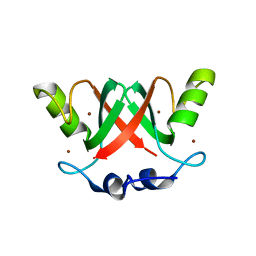 | | Crystal Structure of the MDM2-MDMX RING Domain Heterodimer | | Descriptor: | CITRATE ANION, E3 UBIQUITIN-PROTEIN LIGASE MDM2, MDM4 PROTEIN, ... | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-05-13 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Mdm2/Mdmx Ring Domain Heterodimer Reveals Dimerization is Required for Their Ubiquitylation in Trans.
Cell Death Differ., 15, 2008
|
|
2VJE
 
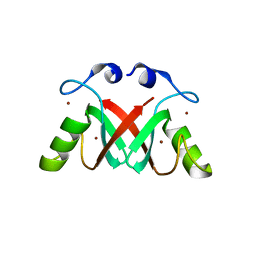 | | Crystal Structure of the MDM2-MDMX RING Domain Heterodimer | | Descriptor: | CITRATE ANION, E3 UBIQUITIN-PROTEIN LIGASE MDM2, MDM4 PROTEIN, ... | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-05-13 | | Last modified: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans.
Cell Death Differ., 15, 2008
|
|
5CEK
 
 | |
5CEM
 
 | |
5ULM
 
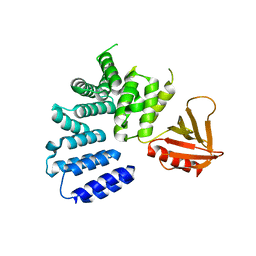 | |
7UPM
 
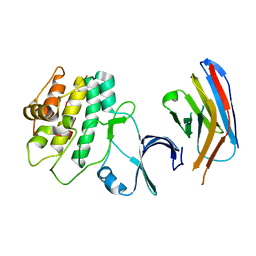 | |
5EDV
 
 | | Structure of the HOIP-RBR/UbcH5B~ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Lechtenberg, B.C, Mace, P.D, Sanishvili, R, Riedl, S.J. | | Deposit date: | 2015-10-22 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation.
Nature, 529, 2016
|
|
4W50
 
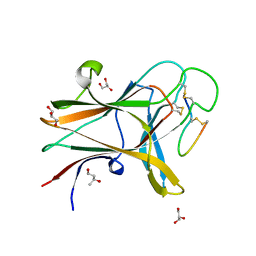 | | Structure of the EphA4 LBD in complex with peptide | | Descriptor: | 1,3-BUTANEDIOL, APY peptide, Ephrin type-A receptor 4, ... | | Authors: | Lechtenberg, B.C, Mace, P.D, Riedl, S.J. | | Deposit date: | 2014-08-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Development and Structural Analysis of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor.
Acs Chem.Biol., 9, 2014
|
|
4W4Z
 
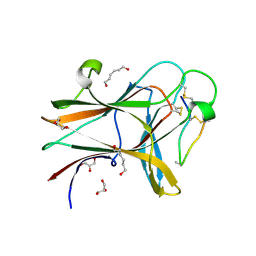 | | Structure of the EphA4 LBD in complex with peptide | | Descriptor: | APY-bAla8.am peptide, Ephrin type-A receptor 4, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Mace, P.D, Riedl, S.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Development and Structural Analysis of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor.
Acs Chem.Biol., 9, 2014
|
|
6CGA
 
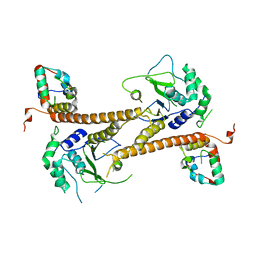 | | Structure of the PR-DUB complex | | Descriptor: | Polycomb protein Asx, Ubiquitin carboxyl-terminal hydrolase calypso | | Authors: | Foglizzo, M, Middleton, A.J, Day, C.L, Mace, P.D. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A bidentate Polycomb Repressive-Deubiquitinase complex is required for efficient activity on nucleosomes.
Nat Commun, 9, 2018
|
|
6DC0
 
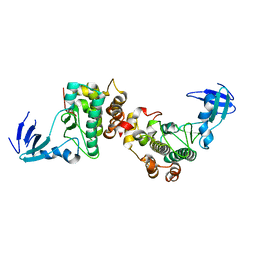 | |
5D0K
 
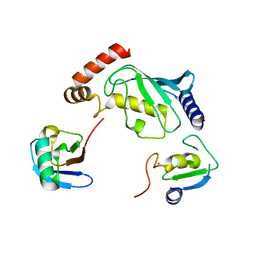 | | Structure of UbE2D2:RNF165:Ub complex | | Descriptor: | Polyubiquitin-B, RING finger protein 165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D0I
 
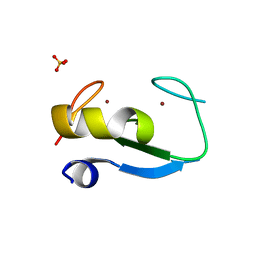 | | Structure of RING finger protein 165 | | Descriptor: | RING finger protein 165, SULFATE ION, ZINC ION | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D0M
 
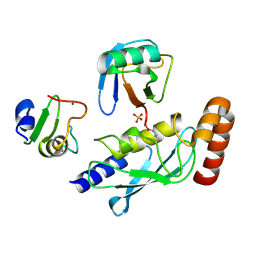 | | Structure of UbE2D2:RNF165:Ub complex | | Descriptor: | PHOSPHATE ION, Polyubiquitin-B, RING finger protein 165, ... | | Authors: | Wright, J.D, Day, C.L, Mace, P.D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-12-09 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6VH9
 
 | | FphF, Staphylococcus aureus fluorophosphonate-binding serine hydrolases F, apo form | | Descriptor: | Esterase family protein, SODIUM ION | | Authors: | Fellner, M, Jamieson, S.A, Brewster, J.L, Mace, P.D. | | Deposit date: | 2020-01-09 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Basis for the Inhibitor and Substrate Specificity of the Unique Fph Serine Hydrolases of Staphylococcus aureus .
Acs Infect Dis., 6, 2020
|
|
