8R11
 
 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R16
 
 | | Structure of compound 12 bound to SARS-CoV-2 main protease | | Descriptor: | 1,2-ETHANEDIOL, 1-[6,7-bis(chloranyl)-3,4-dihydro-1H-isoquinolin-2-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R14
 
 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | Descriptor: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R12
 
 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | Descriptor: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
4PIS
 
 | | Crystal structure of human adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | N~2~-[(2R)-2-(3,5-dichlorophenyl)-2-(dimethylamino)acetyl]-N-({2-[(Z)-iminomethyl]pyrimidin-4-yl}methyl)-L-isoleucinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIE
 
 | | Crystal structure of human adenovirus 2 protease a substrate based nitrile inhibitor | | Descriptor: | ACETATE ION, N-{(2S)-2-(3-chlorophenyl)-2-[(methylsulfonyl)amino]acetyl}-L-phenylalanyl-N-[(2Z)-2-iminoethyl]glycinamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, C, Erbel, C, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PID
 
 | | Crystal structure of human adenovirus 2 protease with a weak pyrimidine nitrile inhibitor | | Descriptor: | ACETATE ION, N-benzyl-2-[(Z)-iminomethyl]pyrimidine-5-carboxamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIQ
 
 | | Crystal structure of human adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | N-[(3,5-dichlorophenyl)acetyl]-L-threonyl-N-[(2Z)-2-iminoethyl]glycinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
1Q0Q
 
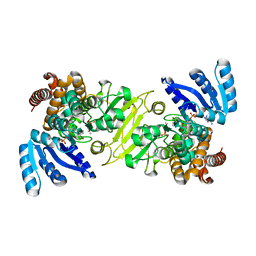 | | Crystal structure of DXR in complex with the substrate 1-deoxy-D-xylulose-5-phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-17 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
1Q0H
 
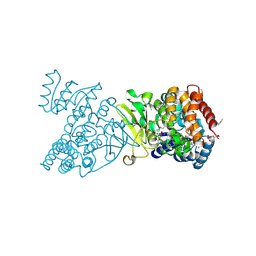 | | Crystal structure of selenomethionine-labelled DXR in complex with fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CITRIC ACID, ... | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
1Q0L
 
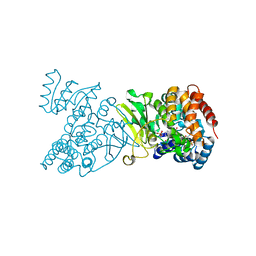 | | Crystal structure of DXR in complex with fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
3EDG
 
 | |
3EDI
 
 | |
3EDH
 
 | |
6G3R
 
 | | Structure of tellurium-centred Anderson-Evans polyoxotungstate (TEW) bound to the nucleotide binding domain of HSP70. Structure one of two TEW-HSP70 structures deposited. | | Descriptor: | 6-tungstotellurate(VI), ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1A, ... | | Authors: | Mac Sweeney, A, Chambovey, A, Wicki, M, Mueller, M, Artico, N, Lange, R, Bijelic, A, Breibeck, J, Rompel, A. | | Deposit date: | 2018-03-26 | | Release date: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystallization additive hexatungstotellurate promotes the crystallization of the HSP70 nucleotide binding domain into two different crystal forms.
PLoS ONE, 13, 2018
|
|
6G3S
 
 | | Structure of tellurium-centred Anderson-Evans polyoxotungstate (TEW) bound to the nucleotide binding domain of HSP70. Second structure of two TEW-HSP70 structures deposited. | | Descriptor: | 6-tungstotellurate(VI), ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1A, ... | | Authors: | Mac Sweeney, A, Chambovey, A, Wicki, M, Mueller, M, Artico, N, Lange, R, Bijelic, A, Breibeck, J, Rompel, A. | | Deposit date: | 2018-03-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystallization additive hexatungstotellurate promotes the crystallization of the HSP70 nucleotide binding domain into two different crystal forms.
PLoS ONE, 13, 2018
|
|
1DLK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF DELTA-CHYMOTRYPSIN BOUND TO A PEPTIDYL CHLOROMETHYL KETONE INHIBITOR | | Descriptor: | CHLORIDE ION, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Mac Sweeney, A, Birrane, G, Walsh, M.A, O'Connell, T, Malthouse, J.P.G. | | Deposit date: | 1999-12-10 | | Release date: | 2000-05-03 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of delta-chymotrypsin bound to a peptidyl chloromethyl ketone inhibitor.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5NB7
 
 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NAR
 
 | |
5NAW
 
 | | Complement factor D in complex with the inhibitor (1R,3S,5R)-2-Aza-bicyclo[3.1.0]hexane-2,3-dicarboxylic acid 2-[(1-carbamoyl-1H-indol-3-yl)-amide] 3-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (1~{R},3~{S},5~{R})-~{N}2-(1-aminocarbonylindol-3-yl)-~{N}3-[3-(trifluoromethyloxy)phenyl]-2-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NBA
 
 | | Complement factor D in complex with the inhibitor (2S,4R)-4-Fluoro-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (2~{S},4~{R})-~{N}1-(1-aminocarbonylindol-3-yl)-4-fluoranyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NB6
 
 | | Complement factor D in complex with the inhibitor (2S,4S)-4-Amino-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (2~{S},4~{S})-~{N}1-(1-aminocarbonylindol-3-yl)-4-azanyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NAT
 
 | |
5FCR
 
 | | MOUSE COMPLEMENT FACTOR D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Complement factor D, DIMETHYL SULFOXIDE, ... | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FCK
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[[(1~{R})-1-(3-chloranyl-2-fluoranyl-phenyl)ethyl]carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]pyrazolo[3,4-c]pyridine-3-carboxamide, Complement factor D, SULFATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
