2OGT
 
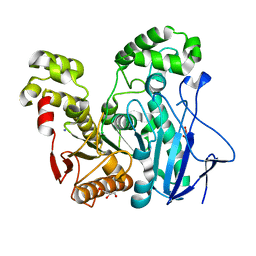 | | Crystal Structure of the Geobacillus Stearothermophilus Carboxylesterase EST55 at pH 6.8 | | Descriptor: | GLYCEROL, IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
2OGS
 
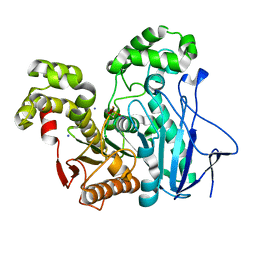 | | Crystal Structure of the GEOBACILLUS STEAROTHERMOPHILUS Carboxylesterase EST55 at pH 6.2 | | Descriptor: | IODIDE ION, Thermostable carboxylesterase Est50 | | Authors: | Liu, P, Ewis, H.E, Tai, P.C, Lu, C.D, Weber, I.T. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est55 and Its Activation of Prodrug CPT-11.
J.Mol.Biol., 367, 2007
|
|
2RCV
 
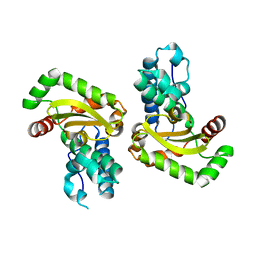 | | Crystal structure of the Bacillus subtilis superoxide dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Liu, P, Ewis, H.E, Huang, Y.J, Lu, C.D, Tai, P.C, Weber, I.T. | | Deposit date: | 2007-09-20 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacillus subtilis superoxide dismutase.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1TQH
 
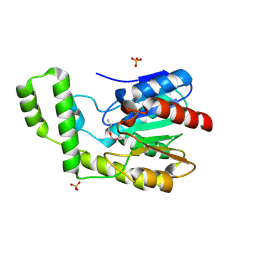 | | Covalent Reaction intermediate Revealed in Crystal Structure of the Geobacillus stearothermophilus Carboxylesterase Est30 | | Descriptor: | Carboxylesterase precursor, PROPYL ACETATE, SULFATE ION | | Authors: | Liu, P, Wang, Y.F, Ewis, H.E, Abdelal, A.T, Lu, C.D, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30.
J.Mol.Biol., 342, 2004
|
|
3NYC
 
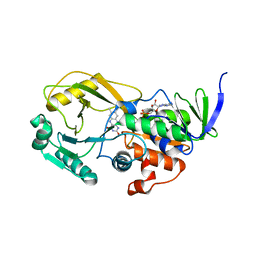 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase | | Descriptor: | (2E)-5-[(diaminomethylidene)amino]-2-iminopentanoic acid, D-Arginine Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fu, G, Weber, I.T. | | Deposit date: | 2010-07-14 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Conformational changes and substrate recognition in Pseudomonas aeruginosa D-arginine dehydrogenase.
Biochemistry, 49, 2010
|
|
3NYF
 
 | |
3NYE
 
 | |
