6YJ1
 
 | | The M23 peptidase domain of the Staphylococcal phage 2638A endolysin | | Descriptor: | ORF007, ZINC ION | | Authors: | Dunne, M, Ernst, P, Sobieraj, A, Pluckthun, A, Loessner, M.J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | (CASP target) Crystal structure of the M23 peptidase domain of Staphylococcal phage 2638A endolysin
To Be Published
|
|
7AQH
 
 | | Cell wall binding domain of the Staphylococcal phage 2638A endolysin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ORF007 | | Authors: | Dunne, M, Sobieraj, A, Ernst, P, Mittl, P.R.E, Pluckthun, A, Loessner, M.J. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Cell wall binding domain of the Staphylococcal phage 2638A endolysin
To Be Published
|
|
6R5W
 
 | | Crystal structure of the receptor binding protein (gp15) of Listeria phage PSA | | Descriptor: | ACETATE ION, CADMIUM ION, Gp15 protein, ... | | Authors: | Dunne, M, Ernst, P, Pluckthun, A, Loessner, M.J, Kilcher, S. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-30 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins.
Cell Rep, 29, 2019
|
|
6F45
 
 | | Crystal structure of the gp37-gp38 adhesin tip complex of the bacteriophage S16 long tail fiber | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, IMIDAZOLE, ... | | Authors: | Dunne, M, Leiman, P, Klumpp, J, Loessner, M.J. | | Deposit date: | 2017-11-29 | | Release date: | 2018-08-01 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (1.70355821 Å) | | Cite: | Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition.
Structure, 26, 2018
|
|
6HX0
 
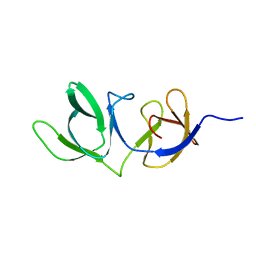 | | Cell wall binding domain of endolysin from Listeria phage A500. | | Descriptor: | L-alanyl-D-glutamate peptidase | | Authors: | Dunne, M, Taylor, N.M.I, Shen, Y, Loessner, M.J, Leiman, P.G. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis for recognition of bacterial cell wall teichoic acid by pseudo-symmetric SH3b-like repeats of a viral peptidoglycan hydrolase
Chem Sci, 2020
|
|
7B1R
 
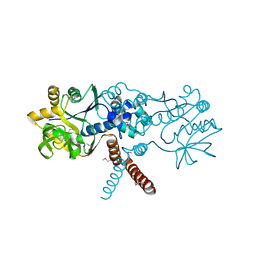 | |
3DKT
 
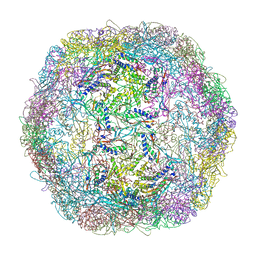 | | Crystal structure of Thermotoga maritima encapsulin | | Descriptor: | Maritimacin, Putative uncharacterized protein | | Authors: | Sutter, M, Boehringer, D, Gutmann, S, Weber-Ban, E, Ban, N. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis of enzyme encapsulation into a bacterial nanocompartment
Nat.Struct.Mol.Biol., 15, 2008
|
|
1XOV
 
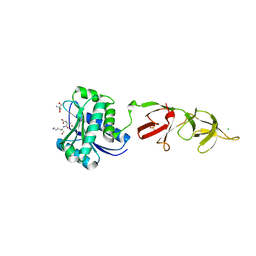 | |
2VO9
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATICALLY ACTIVE DOMAIN OF THE LISTERIA MONOCYTOGENES BACTERIOPHAGE 500 ENDOLYSIN PLY500 | | Descriptor: | L-ALANYL-D-GLUTAMATE PEPTIDASE, SULFATE ION, ZINC ION | | Authors: | Korndoerfer, I.P, Kanitz, A, Skerra, A. | | Deposit date: | 2008-02-09 | | Release date: | 2008-02-19 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of the L-Alanoyl-D-Glutamate Endopeptidase Domain of Listeria Bacteriophage Endolysin Ply500 Reveals a New Member of the Las Peptidase Family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6HHK
 
 | |
