2AIU
 
 | | Crystal Structure of Mouse Testicular Cytochrome C at 1.6 Angstrom | | Descriptor: | Cytochrome c, testis-specific, PHOSPHATE ION, ... | | Authors: | Liu, Z, Ye, S, Lin, H, Rao, Z, Liu, X.J. | | Deposit date: | 2005-08-01 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Remarkably high activities of testicular cytochrome c in destroying reactive oxygen species and in triggering apoptosis
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1D8B
 
 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
1G3F
 
 | | NMR STRUCTURE OF A 9 RESIDUE PEPTIDE FROM SMAC/DIABLO COMPLEXED TO THE BIR3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN 3, SMAC, ZINC ION | | Authors: | Liu, Z, Sun, C, Olejniczak, E.T, Meadows, R.P, Betz, S.F, Oost, T, Herrmann, J, Wu, J.C, Fesik, S.W. | | Deposit date: | 2000-10-24 | | Release date: | 2001-01-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain.
Nature, 408, 2000
|
|
1Y29
 
 | |
2N2K
 
 | | Ensemble structure of the closed state of Lys63-linked diubiquitin in the absence of a ligand | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ubiquitin | | Authors: | Liu, Z, Gong, Z, Tang, C. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-08 | | Last modified: | 2015-09-23 | | Method: | SOLUTION NMR | | Cite: | Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife, 4, 2015
|
|
5KQZ
 
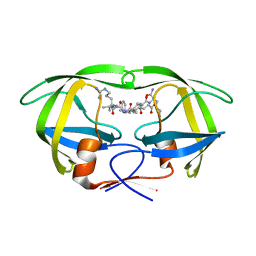 | | Protease E35D-CaP2 | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, Protease | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KQY
 
 | | Protease E35D-DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease E35D-DRV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR1
 
 | | Protease PR5-DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease PR5-DRV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR0
 
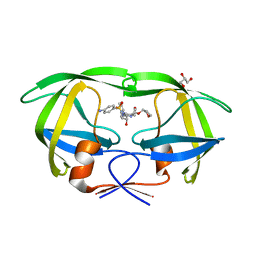 | | Protease E35D-APV | | Descriptor: | GLYCEROL, Protease E35D-APV, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KQX
 
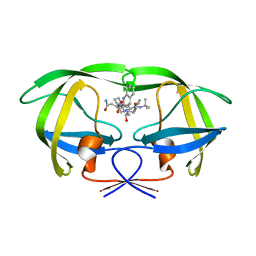 | | Protease E35D-SQV | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease E35D-SQV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR2
 
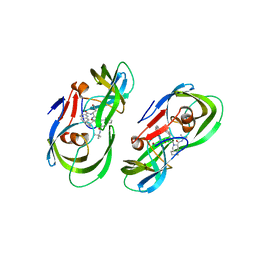 | | Protease PR5-SQV | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease PR5-SQV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
1K1G
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
4R7A
 
 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
1RWT
 
 | | Crystal Structure of Spinach Major Light-harvesting complex at 2.72 Angstrom Resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Liu, Z, Yan, H, Wang, K, Kuang, T, Zhang, J, Gui, L, An, X, Chang, W. | | Deposit date: | 2003-12-17 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of spinach major light-harvesting complex at 2.72 A resolution
Nature, 428, 2004
|
|
6VIE
 
 | | Structure of caspase-1 in complex with gasdermin D | | Descriptor: | Caspase-1 subunit p10, Caspase-1 subunit p20, Gasdermin-D | | Authors: | Liu, Z, Xiao, T.S. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Caspase-1 Engages Full-Length Gasdermin D through Two Distinct Interfaces That Mediate Caspase Recruitment and Substrate Cleavage.
Immunity, 53, 2020
|
|
3HNB
 
 | |
3HOB
 
 | |
3HNY
 
 | |
3JCI
 
 | | 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles | | Descriptor: | Capsid protein | | Authors: | Liu, Z, Guo, F, Wang, F, Li, T.C, Jiang, W. | | Deposit date: | 2015-12-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | 2.9 angstrom Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles.
Structure, 24, 2016
|
|
3LFK
 
 | | A reported archaeal mechanosensitive channel is a structural homolog of MarR-like transcriptional regulators | | Descriptor: | CITRIC ACID, MarR Like Protein, TVG0766549, ... | | Authors: | Liu, Z, Walton, T.A, Rees, D.C. | | Deposit date: | 2010-01-17 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A reported archaeal mechanosensitive channel is a structural homolog of MarR-like transcriptional regulators.
Protein Sci., 19, 2010
|
|
2JIX
 
 | | Crystal structure of ABT-007 FAB fragment with the soluble domain of EPO receptor | | Descriptor: | ABT-007 FAB FRAGMENT, ERYTHROPOIETIN RECEPTOR | | Authors: | Liu, Z, Stoll, V.S, DeVries, P, Jakob, C.G, Xie, N, Simmer, R.L, Lacy, S.E, Egan, D.A, Harlan, J.E, Lesniewski, R.R, Reilly, E.B. | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-10 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Potent Erythropoietin-Mimicking Human Antibody Interacts Through a Novel Binding Site.
Blood, 110, 2007
|
|
2LQC
 
 | |
2LQP
 
 | |
8WU2
 
 | | Crystal structure of RNA-dependent RNA polymerases from Jingmen tick virus | | Descriptor: | RNA polymerase, SELENIUM ATOM | | Authors: | Liu, Z, Han, P, Peng, Q, Qi, J. | | Deposit date: | 2023-10-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RNA-dependent RNA polymerases from Jingmen tick virus and Alongshan virus
HLIFE, 2, 2024
|
|
8WA8
 
 | | Human transketolase in complex with phosphite | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHITE ION, ... | | Authors: | Liu, Z, Tittmann, K, Dai, S. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
