1TI5
 
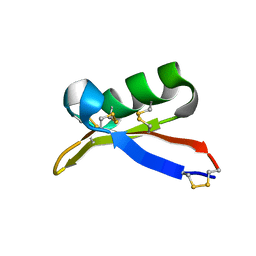 | | Solution structure of plant defensin | | Descriptor: | plant defensin | | Authors: | Liu, Y.J, Cheng, C.S, Liu, Y.N, Hsu, M.P, Chen, C.S, Lyu, P.C. | | Deposit date: | 2004-06-02 | | Release date: | 2005-07-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids.
Proteins, 63, 2006
|
|
5YJ6
 
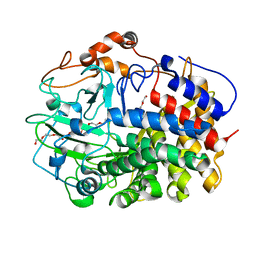 | | The exoglucanase CelS from Clostridium thermocellum | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, Dockerin type I repeat-containing protein | | Authors: | Liu, Y.J, Liu, S.Y, Dong, S, Li, R.M, Feng, Y.G, Cui, Q. | | Deposit date: | 2017-10-09 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Determination of the native features of the exoglucanase Cel48S from Clostridium thermocellum
Biotechnol Biofuels, 11, 2018
|
|
5XZ4
 
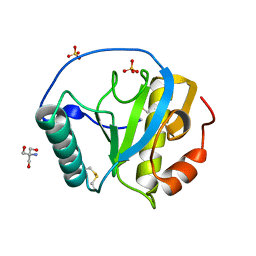 | | The X-tay structure of Bumblebee PGRP-SA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bumblebee peptidoglycan recognition protein SA, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Insights into the Preferential Binding of PGRP-SAs from Bumblebees and Honeybees to Dap-Type Peptidoglycans Rather than Lys-Type Peptidoglycans.
J Immunol., 202, 2019
|
|
5YJ7
 
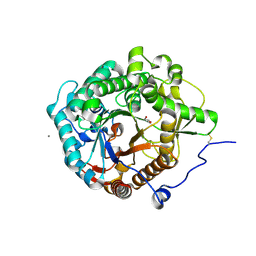 | | Structural insight into the beta-GH1 glucosidase BGLN1 from oleaginous microalgae Nannochloropsis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase | | Authors: | Dong, S, Liu, Y.J, Zhou, H.X, Xiao, Y, Xu, J, Cui, Q, Wang, X.Q, Feng, Y.G. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insight into a GH1 beta-glucosidase from the oleaginous microalga, Nannochloropsis oceanica.
Int.J.Biol.Macromol., 170, 2021
|
|
5NS3
 
 | |
5NS4
 
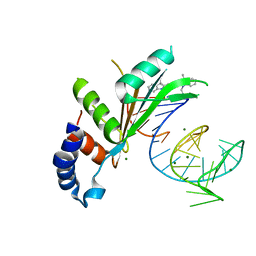 | | Crystal structures of Cy3 cyanine fluorophores stacked onto the end of double-stranded RNA | | Descriptor: | 3-[(2~{Z})-2-[(~{E})-3-[3,3-dimethyl-1-(3-oxidanylpropyl)indol-1-ium-2-yl]prop-2-enylidene]-3,3-dimethyl-indol-1-yl]propan-1-ol, 50S ribosomal protein L5, MAGNESIUM ION, ... | | Authors: | Liu, Y.J, Lilley, D.M.J. | | Deposit date: | 2017-04-25 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Cyanine Fluorophores Stacked onto the End of Double-Stranded RNA.
Biophys. J., 113, 2017
|
|
5CNQ
 
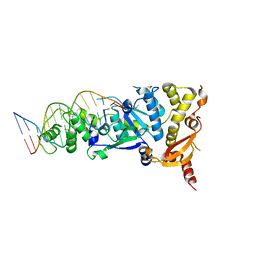 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
5CO8
 
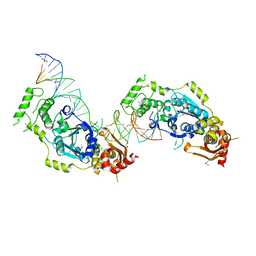 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA and Mg2+ ion | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*GP*AP*CP*TP*GP*CP*AP*GP*TP*TP*GP*AP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
5EB9
 
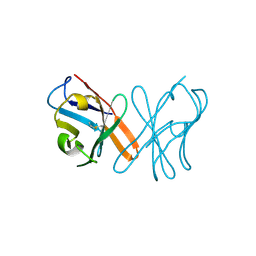 | | Crystal Structure Of Chicken CD8aa Homodimer | | Descriptor: | CD8 alpha chain | | Authors: | Liu, Y.J, Qi, J.X, Xia, C. | | Deposit date: | 2015-10-18 | | Release date: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
5EDX
 
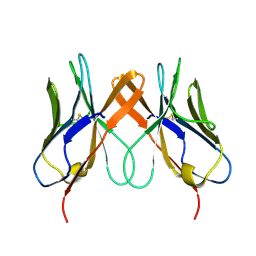 | | Crystal structure of swine CD8aa homodimer | | Descriptor: | CD8 alpha antigen | | Authors: | Liu, Y.J, Qi, J.X, Zhang, N.Z, Xia, C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
5XZ3
 
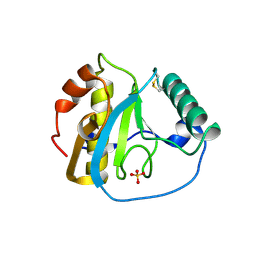 | | The X-ray structure of Apis mellifera PGRP-SA | | Descriptor: | Peptidoglycan-recognition protein, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae).
Protein Sci., 27, 2018
|
|
7BWN
 
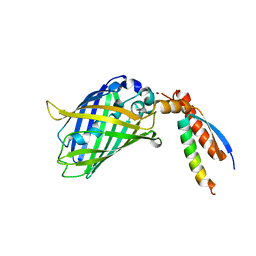 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LHH
 
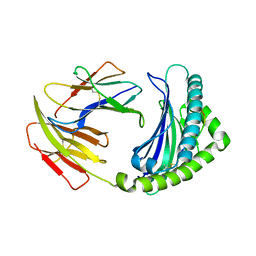 | | Crystal structure of chicken 8mer-BF2*1501 | | Descriptor: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LHF
 
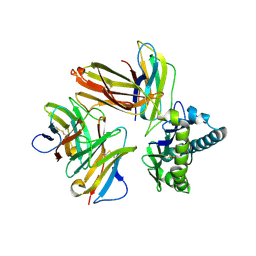 | | Crystal structure of chicken cCD8aa/pBF2*15:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, MHC class I, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LHG
 
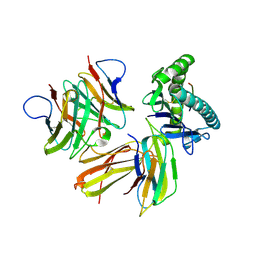 | | Crystal structure of chicken cCD8aa/pBF2*04:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, IE8 peptide, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
1L6H
 
 | |
5H1Y
 
 | |
8UD9
 
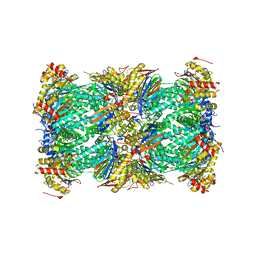 | | Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8X39
 
 | |
8X3A
 
 | |
8G6E
 
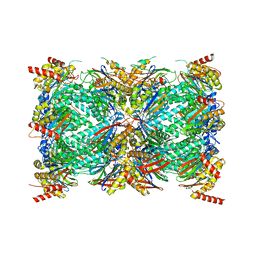 | | Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8G6F
 
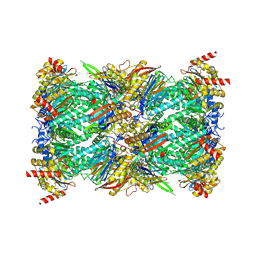 | | Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, Proteasome endopeptidase complex, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
4Y29
 
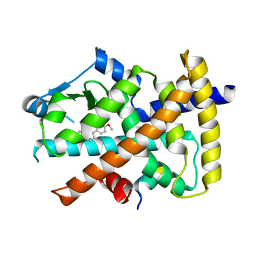 | | Identification of a novel PPARg ligand that regulates metabolism | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, R, Li, Y. | | Deposit date: | 2015-02-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective targeting of PPAR gamma by the natural product chelerythrine with a unique binding mode and improved antidiabetic potency.
Sci Rep, 5, 2015
|
|
6IVS
 
 | |
6IVU
 
 | |
