1IU9
 
 | | Crystal structure of the C-terminal domain of aspartate racemase from Pyrococcus horikoshii OT3 | | Descriptor: | CALCIUM ION, aspartate racemase | | Authors: | Liu, L, Iwata, K, Yohda, M, Miki, K. | | Deposit date: | 2002-02-28 | | Release date: | 2003-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insight into gene duplication, gene fusion and domain swapping in the evolution of PLP-independent amino acid racemases
FEBS LETT., 528, 2002
|
|
1IUA
 
 | | Ultra-high resolution structure of HiPIP from Thermochromatium tepidum | | Descriptor: | High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Liu, L, Nogi, T, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2002-03-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ultrahigh-resolution structure of high-potential iron-sulfur protein from Thermochromatium tepidum.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5WYB
 
 | | Structure of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
3HH3
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - 1,2-dihydro-1,2-azaborine | | Descriptor: | 1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH4
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - Benzene as control | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BENZENE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH5
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - 1-ethyl-2-hydro-1,2-azaborine | | Descriptor: | 1-ethyl-1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HH6
 
 | | New azaborine compounds bind to the T4 lysozyme L99A cavity -ethylbenzene as control | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, PHENYLETHANE, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1T2L
 
 | | Three Crystal Structures of Human Coactosin-like Protein | | Descriptor: | Coactosin-like protein | | Authors: | Liu, L, Wei, Z, Chen, Z, Wang, Y, Gong, W. | | Deposit date: | 2004-04-22 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Coactosin-like Protein
J.Mol.Biol., 344, 2004
|
|
1T3X
 
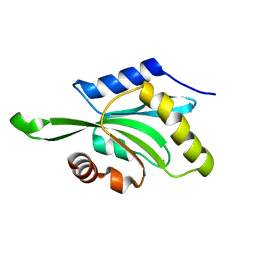 | |
1T3Y
 
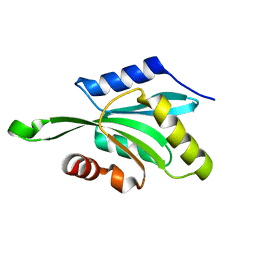 | |
4YL9
 
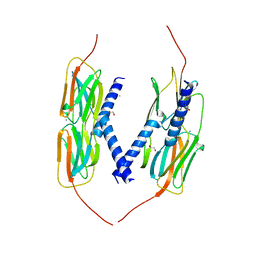 | | Crystal Structure of wild-type of hsp14.1 from Sulfolobus solfatataricus P2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heat shock protein Hsp20 | | Authors: | Liu, L, Chen, J.Y, Yun, C.H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Active-State Structures of a Small Heat-Shock Protein Revealed a Molecular Switch for Chaperone Function
Structure, 23, 2015
|
|
4YLC
 
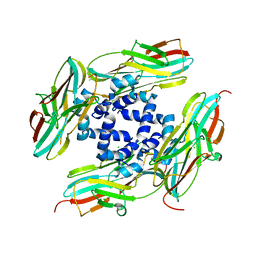 | |
4YLB
 
 | |
8HE4
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
 | |
8HE1
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HF9
 
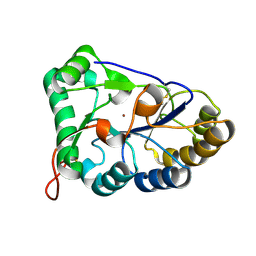 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
2B3R
 
 | | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2 | | Descriptor: | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide, SULFATE ION | | Authors: | Liu, L, Song, X, He, D, Komma, C, Kita, A, Verbasius, J.V, Bellamy, H, Miki, K, Czech, M.P, Zhou, G.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2alpha.
J.Biol.Chem., 281, 2006
|
|
5WYD
 
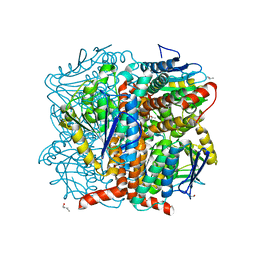 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
3CIG
 
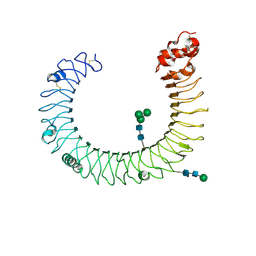 | | Crystal structure of mouse TLR3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
3CIY
 
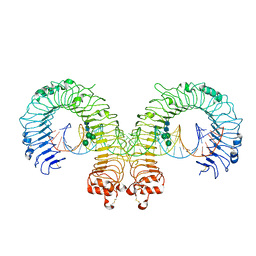 | | Mouse Toll-like receptor 3 ectodomain complexed with double-stranded RNA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
3DN1
 
 | | Chloropentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1-chloro-2,3,4,5,6-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
3DMZ
 
 | |
3DMV
 
 | |
