4LQI
 
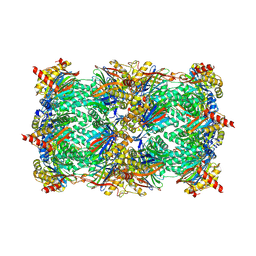 | | Yeast 20S Proteasome in complex with Vibralactone | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | List, A, Zeiler, E, Gallastegui, N, Rusch, M, Hedberg, C, Sieber, S.A, Groll, M. | | Deposit date: | 2013-07-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Omuralide and Vibralactone: Differences in the Proteasome-beta-Lactone-gamma-Lactam Binding Scaffold Alter Target Preferences.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3V5I
 
 | |
4FFO
 
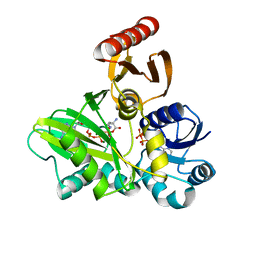 | | PylC in complex with phosphorylated D-ornithine | | Descriptor: | (2R)-2,5-diaminopentanoyl dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
4FFP
 
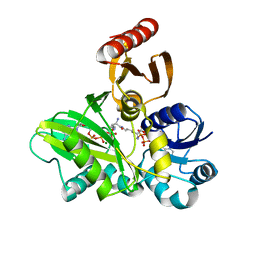 | | PylC in complex with L-lysine-Ne-D-ornithine (cocrystallized with L-lysine and D-ornithine) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N~6~-D-ornithyl-L-lysine, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
4FFL
 
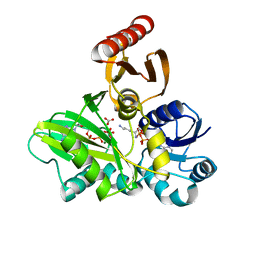 | | PylC in complex with L-lysine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
4FFN
 
 | | PylC in complex with D-ornithine and AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ORNITHINE, MAGNESIUM ION, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
4FFM
 
 | | PylC in complex with L-lysine-Ne-D-ornithine (cocrystallized with L-lysine-Ne-D-ornithine) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
4FFR
 
 | | SeMet-labeled PylC (remote) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PylC | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
3T7V
 
 | | Crystal structure of methylornithine synthase (PylB) | | Descriptor: | 5-amino-D-isoleucine, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Quitterer, F, List, A, Eisenreich, W, Bacher, A, Groll, M. | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4J70
 
 | | Yeast 20S proteasome in complex with the belactosin derivative 3e | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Kawamura, S, Unno, Y, List, A, Tanaka, M, Sasaki, T, Arisawa, M, Asai, A, Groll, M, Shuto, S. | | Deposit date: | 2013-02-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent Proteasome Inhibitors Derived from the Unnatural cis-Cyclopropane Isomer of Belactosin A: Synthesis, Biological Activity, and Mode of Action.
J.Med.Chem., 56, 2013
|
|
3V5E
 
 | | Crystal structure of ClpP from Staphylococcus aureus in the active, extended conformation | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Gersch, M, List, A, Groll, M, Sieber, S. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into structural network responsible for oligomerization and activity of bacterial virulence regulator caseinolytic protease P (ClpP) protein.
J.Biol.Chem., 287, 2012
|
|
4JCR
 
 | | ClpP1 N165D mutant from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zeiler, E, List, A, Alte, F, Gersch, M, Wachtel, R, Groll, M, Sieber, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JCQ
 
 | | ClpP1 from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zeiler, E, List, A, Alte, F, Gersch, M, Wachtel, R, Groll, M, Sieber, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JCT
 
 | | ClpP2 from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zeiler, E, List, A, Alte, F, Gersch, M, Wachtel, R, Groll, M, Sieber, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
5VZ2
 
 | |
6CFD
 
 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
