2YIZ
 
 | | X-ray structure of Mycobacterium tuberculosis Dodecin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Vinzenz, X, Grosse, W, Linne, U, Meissner, B, Essen, L.-O. | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Engineering of Mycobacterium Tuberculosis Dodecin Hybrids.
Chem.Commun.(Camb.), 47, 2011
|
|
2YJ0
 
 | | X-ray structure of chemically engineered Mycobacterium tuberculosis Dodecin | | Descriptor: | CHLORIDE ION, COENZYME A, DODECIN, ... | | Authors: | Vinzenz, X, Grosse, W, Linne, U, Meissner, B, Essen, L.-O. | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical Engineering of Mycobacterium Tuberculosis Dodecin Hybrids.
Chem.Commun.(Camb.), 47, 2011
|
|
2MMW
 
 | |
5A8J
 
 | | Crystal structure of the ArnB paralog VWA2 from Sulfolobus acidocaldarius | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Hoffmann, L, Anders, K, Reimann, J, Linne, U, Essen, L.-O, Albers, S.-V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Phosphorylation-Dependent Interaction between an Archaeal Von Willebrand and Fha Domain Recruits Arna-Arnb Complex to DNA for Repression of Motility
To be Published
|
|
5A8I
 
 | | Crystal structure of the FHA domain of ArnA from Sulfolobus acidocaldarius | | Descriptor: | ARNA, GLYCEROL, SULFATE ION | | Authors: | Hoffmann, L, Anders, K, Reimann, J, Linne, U, Essen, L.-O, Albers, S.-V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphorylation-Dependent Interaction between an Archaeal Von Willebrand and Fha Domain Recruits Arna-Arnb Complex to DNA for Repression of Motility
To be Published
|
|
3FYX
 
 | | The Structure of OmpF porin with a synthetic dibenzo-18-crown-6 as modulator | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(6,7,9,10,17,18,20,21-octahydrodibenzo[b,k][1,4,7,10,13,16]hexaoxacyclooctadecin-2-yl)acetamide, Outer membrane protein F | | Authors: | Reitz, S, Cebi, M, Koert, U, Essen, L.O. | | Deposit date: | 2009-01-23 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | On the function and structure of synthetically modified porins.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4RZ2
 
 | |
4RZ3
 
 | | Crystal structure of the MinD-like ATPase FlhG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Site-determining protein | | Authors: | Schuhmacher, J.S, Bange, G. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3KIG
 
 | | Mutant carbonic anhydrase II in complex with an azide and an alkyne | | Descriptor: | 2-azido-N-(2-sulfanylethyl)ethanamide, 3-ethynylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Schulze-Wischeler, J, Niehage, N.U, Heine, A, Klebe, G. | | Deposit date: | 2009-11-02 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Stereo- and Regioselective Azide/Alkyne Cycloadditions in Carbonic Anhydrase II via Tethering, Monitored by Crystallography and Mass Spectrometry.
Chemistry, 17, 2011
|
|
3KNE
 
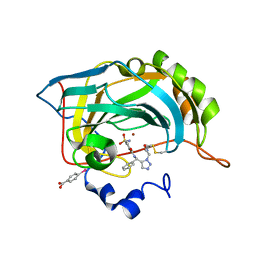 | | Carbonic Anhydrase II H64C mutant in complex with an in situ formed triazole | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, N-[(S)-(1-{2-oxo-2-[(3-sulfanylpropyl)amino]ethyl}-1H-1,2,3-triazol-5-yl)(phenyl)methyl]-4-sulfamoylbenzamide, ... | | Authors: | Schulze-Wischeler, J, Heine, A, Klebe, G. | | Deposit date: | 2009-11-12 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Stereo- and Regioselective Azide/Alkyne Cycloadditions in Carbonic Anhydrase II via Tethering, Monitored by Crystallography and Mass Spectrometry.
Chemistry, 17, 2011
|
|
2MJD
 
 | | Oxidized Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
2MJE
 
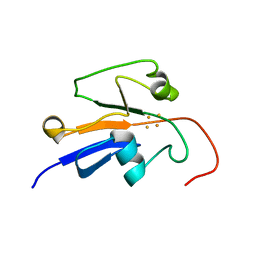 | | Reduced Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
2NAR
 
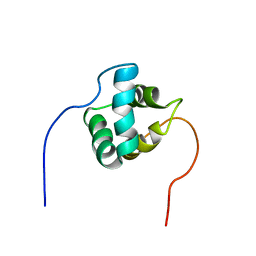 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2018-06-06 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
6GOW
 
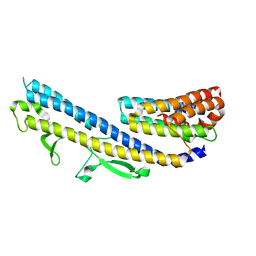 | |
3M2Z
 
 | |
5DED
 
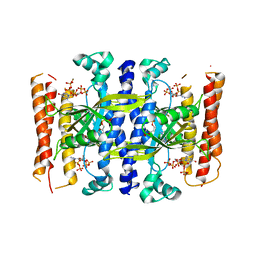 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to its product pppGpp | | Descriptor: | GTP pyrophosphokinase YjbM, MAGNESIUM ION, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5F2V
 
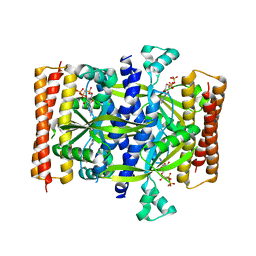 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GTP pyrophosphokinase YjbM, MAGNESIUM ION | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3M5T
 
 | |
5J8Q
 
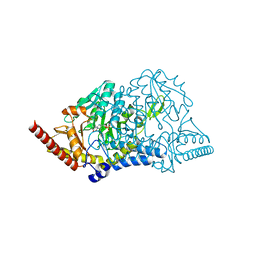 | |
5JRK
 
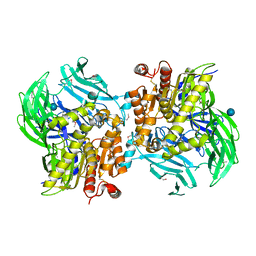 | | Crystal Structure of the Sphingopyxin I Lasso Peptide Isopeptidase SpI-IsoP (SeMet-derived) | | Descriptor: | Dipeptidyl aminopeptidases/acylaminoacyl-peptidases-like protein, beta-D-glucopyranose | | Authors: | Fage, C.D, Hegemann, J.D, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
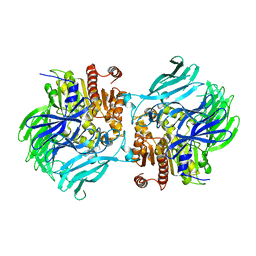
5JRL
 
 | |
5JQF
 
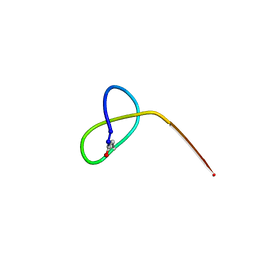 | | Crystal structure of the lasso peptide Sphingopyxin I (SpI) | | Descriptor: | Sphingopyxin I | | Authors: | Fage, C.D, Hegemann, J.D, Harms, K, Bange, G, Marahiel, M.A. | | Deposit date: | 2016-05-04 | | Release date: | 2016-09-14 | | Last modified: | 2021-06-16 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure and Mechanism of the Sphingopyxin I Lasso Peptide Isopeptidase.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5DEC
 
 | |
5LWR
 
 | | Endothiapepsin in complex with a derivative of fragment 177 | | Descriptor: | 1,2-ETHANEDIOL, 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LWS
 
 | | Endothiapepsin in complex with fragment 177 and a derivative thereof | | Descriptor: | 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, 4-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazine, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
