4Y20
 
 | | Complex of human Galectin-1 and NeuAcalpha2-3Galbeta1-4Glc | | Descriptor: | Galectin-1, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1X
 
 | | Complex of human Galectin-1 and Galbeta1-4(6OSO3)GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1V
 
 | | Complex of human Galectin-1 and Galbeta1-3GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1U
 
 | | Complex of human Galectin-1 and Galbeta1-4GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y26
 
 | | Complex of human Galectin-7 and Galbeta1-3(6OSO3)GlcNAc | | Descriptor: | Galectin-7, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1Z
 
 | | Complex of human Galectin-1 and Galbeta1-4(6CO2)GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranosiduronic acid | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y22
 
 | | Complex of human Galectin-1 and (3OSO3)Galbeta1-3GlcNAc | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Galectin-1 | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1Y
 
 | | Complex of human Galectin-1 and (6OSO3)Galbeta1-3GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
7EZQ
 
 | | Complex structure of AtHPPD with inhibitor Y15832 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,5-dimethyl-3-(2-methylphenyl)quinazoline-2,4-dione, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Complex structure of AtHPPD with inhibitor Y18093
To Be Published
|
|
5YWG
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with Mesotrione | | Descriptor: | 2-[(4-methylsulfonyl-2-nitro-phenyl)-oxidanyl-methylidene]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, W.C. | | Deposit date: | 2017-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
7CJK
 
 | | Complex structure of AtHPPD with inhibitor Y18093 | | Descriptor: | 3-oxidanyl-2-[3,5,6-tris(chloranyl)-4-[(4-chlorophenyl)methylamino]pyridin-2-yl]carbonyl-cyclohex-2-en-1-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2020-07-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Complex structure of AtHPPD with inhibitor Y18093
To Be Published
|
|
7E0X
 
 | |
4Y24
 
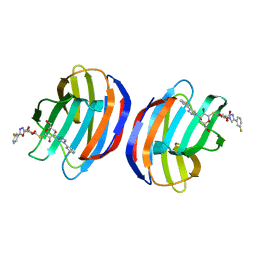 | | Complex of human Galectin-1 and TD-139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-1 | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors.
Sci Rep, 6, 2016
|
|
7V6X
 
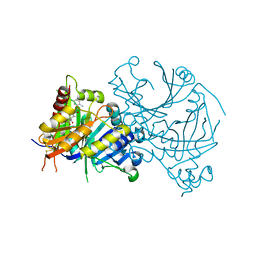 | | Crystal structure of HPPD complexed with Y18556 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-3-[(2-methylphenyl)methyl]-6-[(1~{R},2~{S})-2-oxidanyl-6-oxidanylidene-cyclohexyl]carbonyl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pharmacophore-Oriented Discovery of Novel 1,2,3-Benzotriazine-4-one Derivatives as Potent 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 70, 2022
|
|
5XGK
 
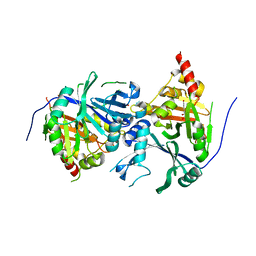 | | Crystal structure of Arabidopsis thaliana 4-hydroxyphenylpyruvate dioxygenase (AtHPPD) complexed with its substrate 4-hydroxyphenylpyruvate acid (HPPA) | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, 4-hydroxyphenylpyruvate dioxygenase, ACETATE ION, ... | | Authors: | Yang, G.F, Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-04-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 4-Hydroxyphenylpyruvate Dioxygenase in Complex with Substrate Reveals a New Starting Point for Herbicide Discovery.
Res, 2019, 2019
|
|
7CQS
 
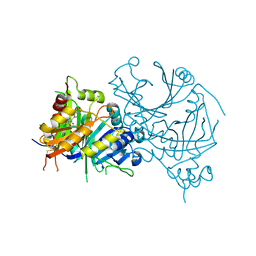 | | Complex structure of HPPD with Topramezone | | Descriptor: | 4-[3-(4,5-dihydro-1,2-oxazol-3-yl)-2-methyl-4-methylsulfonyl-phenyl]carbonyl-2-methyl-1~{H}-pyrazol-3-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2020-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7CQR
 
 | | Complex structure of HPPD with Y16550 | | Descriptor: | 3-(3-chlorophenyl)-6-[(2,5-dimethyl-3-oxidanylidene-1~{H}-pyrazol-4-yl)carbonyl]-1,5-dimethyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2020-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
6JX9
 
 | | Structure of Y17107 complexed HPPD | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-[(~{Z})-(1,3-dimethyl-5-oxidanylidene-pyrazol-4-ylidene)-oxidanyl-methyl]-2-(phenylmethyl)isoindole-1,3-dione, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2019-04-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pyrazole-Isoindoline-1,3-dione Hybrid: A Promising Scaffold for 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 67, 2019
|
|
6LGT
 
 | | Complex structure of HPPD with an inhibitor Y16542 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chlorophenyl)-6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,5-dimethyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.Y, Yang, W.C, Yang, G.F. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Discovery of Novel Pyrazole-Quinazoline-2,4-dione Hybrids as 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 68, 2020
|
|
4XBL
 
 | |
5H9R
 
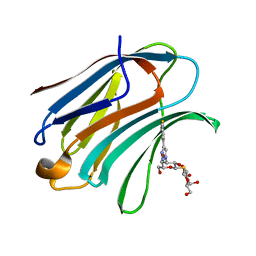 | | Crystal Structure of Human Galectin-3 CRD in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5H9P
 
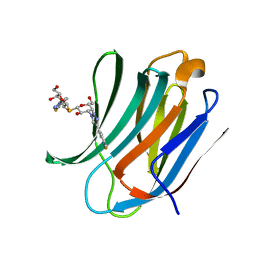 | | Crystal Structure of Human Galectin-3 CRD in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5H9S
 
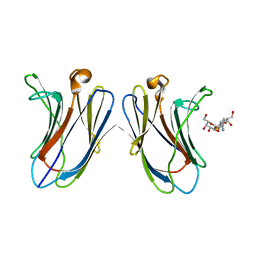 | | Crystal Structure of Human Galectin-7 in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
5H9Q
 
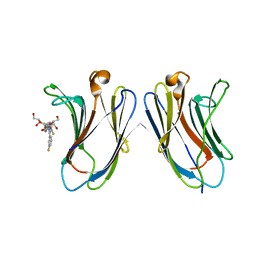 | | Crystal Structure of Human Galectin-7 in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
4XBN
 
 | |
