2N2C
 
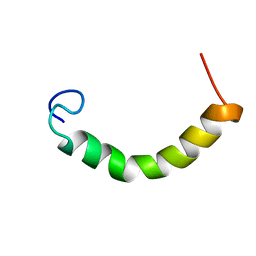 | | NMR Structure of TDP-43 prion-like hydrophobic helix in DPC | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Lim, L, Song, J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | ALS-causing mutations significantly perturb the self-assembly and interaction with nucleic acid of the intrinsically-disordered prion-like domain of TDP-43
To be Published
|
|
2MP3
 
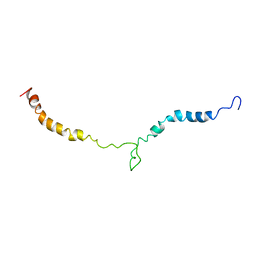 | | Truncated L126Z-sod1 in DPC micelle | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Lim, L, Song, J. | | Deposit date: | 2014-05-10 | | Release date: | 2015-05-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mechanism for transforming cytosolic SOD1 into integral membrane proteins of organelles by ALS-causing mutations
Biochim.Biophys.Acta, 1848, 2015
|
|
2NAM
 
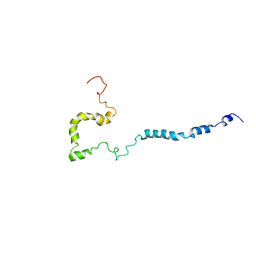 | | Full-length WT SOD1 in DPC MICELLE | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Lim, L, Song, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-12-14 | | Last modified: | 2019-10-16 | | Method: | SOLUTION NMR | | Cite: | SALS-linked WT-SOD1 adopts a highly similar helical conformation as FALS-causing L126Z-SOD1 in a membrane environment
Biochim.Biophys.Acta, 1858, 2016
|
|
2I0K
 
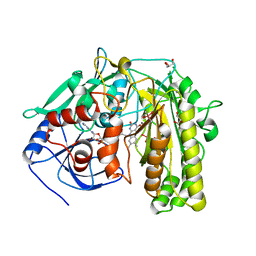 | |
8ESI
 
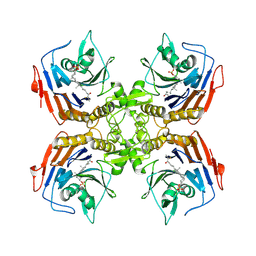 | | Bile Salt Hydrolase from B. longum with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Conjugated bile acid hydrolase | | Authors: | Walker, M.E, Lim, L, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
1BF4
 
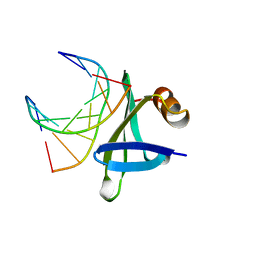 | | CHROMOSOMAL DNA-BINDING PROTEIN SSO7D/D(GCGAACGC) COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*5IUP*CP*GP*C)-3'), PROTEIN (CHROMOSOMAL PROTEIN SSO7D) | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Padmanabhan, S, Lim, L, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
1BNZ
 
 | | SSO7D HYPERTHERMOPHILE PROTEIN/DNA COMPLEX | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-BINDING PROTEIN 7A | | Authors: | Gao, Y.-G, Su, S.-Y, Robinson, H, Padmanabhan, S, Lim, L, Mccrary, B.S, Edmondos, S.P, Shrive, J.W, Wang, A.H.-J. | | Deposit date: | 1998-07-31 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
1CF4
 
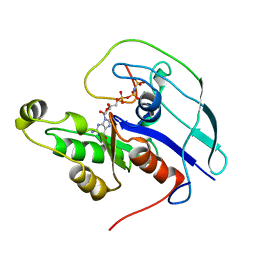 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
2MDK
 
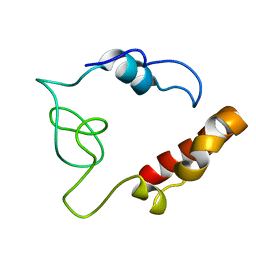 | |
2MBE
 
 | |
6U7J
 
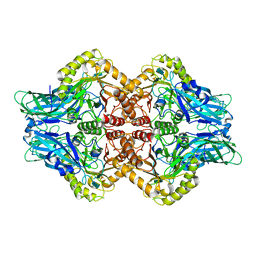 | | Uncultured Clostridium sp. Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CALCIUM ION | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut microbial beta-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens.
J.Biol.Chem., 294, 2019
|
|
2Z2S
 
 | |
6U7I
 
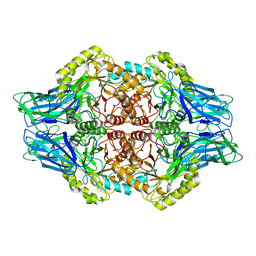 | | Faecalibacterium prausnitzii Beta-glucuronidase | | Descriptor: | Beta-glucuronidase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gut microbial beta-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens.
J.Biol.Chem., 294, 2019
|
|
6USS
 
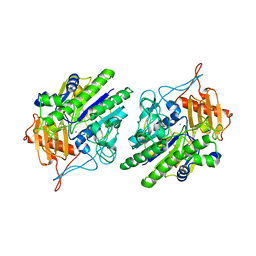 | |
6UST
 
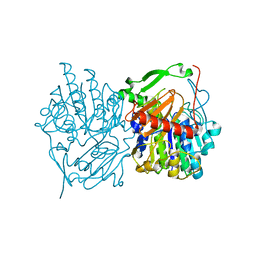 | | Gut microbial sulfatase from Hungatella hathewayi | | Descriptor: | CALCIUM ION, N-acetylgalactosamine 6-sulfate sulfatase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases.
Biochemistry, 59, 2020
|
|
6D7J
 
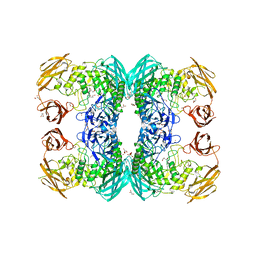 | |
4L67
 
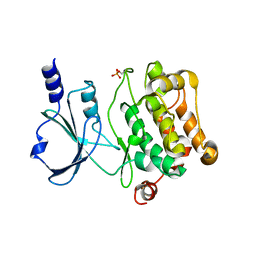 | | Crystal Structure of Catalytic Domain of PAK4 | | Descriptor: | Serine/threonine-protein kinase PAK 4 | | Authors: | Wang, W, Song, J. | | Deposit date: | 2013-06-12 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NMR binding and crystal structure reveal that intrinsically-unstructured regulatory domain auto-inhibits PAK4 by a mechanism different for that of PAK1
Biochem.Biophys.Res.Commun., 438, 2013
|
|
2Q1Z
 
 | |
1CA5
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
1CA6
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
2LW8
 
 | |
