4V7N
 
 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
1GNE
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE OF SCHISTOSOMA JAPONICUM FUSED WITH A CONSERVED NEUTRALIZING EPITOPE ON GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Lim, K, Ho, J.X, Keeling, K, Gilliland, G.L, Ji, X, Ruker, F, Carter, D.C. | | Deposit date: | 1994-06-16 | | Release date: | 1994-11-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV.
Protein Sci., 3, 1994
|
|
1AZF
 
 | | CHICKEN EGG WHITE LYSOZYME CRYSTAL GROWN IN BROMIDE SOLUTION | | Descriptor: | BROMIDE ION, LYSOZYME | | Authors: | Lim, K, Nadarajah, A, Forsythe, E.L, Pusey, M.L. | | Deposit date: | 1997-11-17 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locations of bromide ions in tetragonal lysozyme crystals.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3L2D
 
 | |
3L2E
 
 | | Glycocyamine kinase, alpha-beta heterodimer from marine worm Namalycastis sp. | | Descriptor: | Glycocyamine kinase alpha chain, Glycocyamine kinase beta chain | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
2R82
 
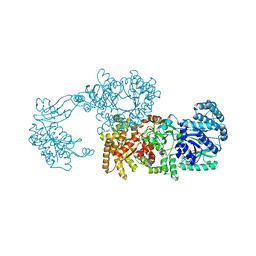 | | Pyruvate phosphate dikinase (PPDK) triple mutant R219E/E271R/S262D adapts a second conformational state | | Descriptor: | Pyruvate, phosphate dikinase, SULFATE ION | | Authors: | Lim, K, Read, R.J, Chen, C.C, Herzberg, O. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Swiveling domain mechanism in pyruvate phosphate dikinase.
Biochemistry, 46, 2007
|
|
1X6J
 
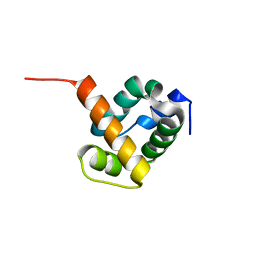 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
1X6I
 
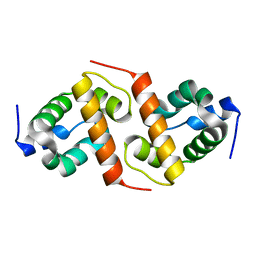 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
1IM8
 
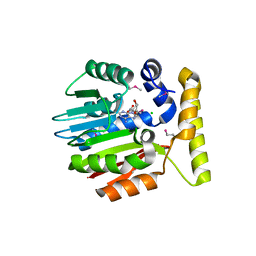 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
1J7G
 
 | |
1J8B
 
 | | Structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function | | Descriptor: | YbaB | | Authors: | Lim, K, Tempcyzk, A, Toedt, J, Parsons, J.F, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-21 | | Release date: | 2003-01-14 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of YbaB from Haemophilus influenzae (HI0442), a
protein of unknown function coexpressed with the recombinational
DNA repair protein RecR
Proteins, 50, 2003
|
|
1J85
 
 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
1MW5
 
 | | Structure of HI1480 from Haemophilus influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI1480 | | Authors: | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-27 | | Release date: | 2003-11-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1MXI
 
 | | Structure of YibK from Haemophilus influenzae (HI0766): a Methyltransferase with a Cofactor Bound at a Site Formed by a Knot | | Descriptor: | Hypothetical tRNA/rRNA methyltransferase HI0766, IODIDE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-02 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a Cofactor Bound at a Site Formed by a Knot
Proteins, 51, 2003
|
|
2PCP
 
 | | ANTIBODY FAB COMPLEXED WITH PHENCYCLIDINE | | Descriptor: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, IMMUNOGLOBULIN | | Authors: | Lim, K, Owens, S.M, Arnold, L, Sacchettini, J.C, Linthicum, D.S. | | Deposit date: | 1998-06-02 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of monoclonal 6B5 Fab complexed with phencyclidine.
J.Biol.Chem., 273, 1998
|
|
4JZ9
 
 | |
4JZ8
 
 | |
4JZ7
 
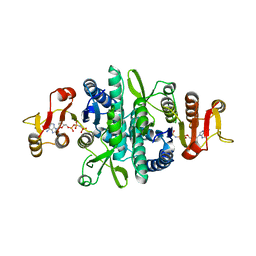 | | Carbamate kinase from Giardia lamblia bound to AMP-PNP | | Descriptor: | Carbamate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lim, K, Herzberg, O. | | Deposit date: | 2013-04-02 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Carbamate Kinase from Giardia lamblia Bound with Citric Acid and AMP-PNP.
Plos One, 8, 2013
|
|
3CA8
 
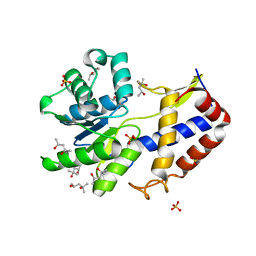 | | Crystal structure of Escherichia coli YdcF, an S-adenosyl-L-methionine utilizing enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein ydcF, SULFATE ION | | Authors: | Lim, K, Chao, K, Lehmann, C, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Escherichia coli YdcF binds S-adenosyl-L-methionine and adopts an alpha/beta-fold characteristic of nucleotide-utilizing enzymes.
Proteins, 72, 2008
|
|
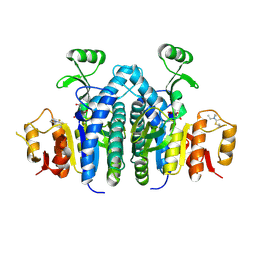
4OLC
 
 | |
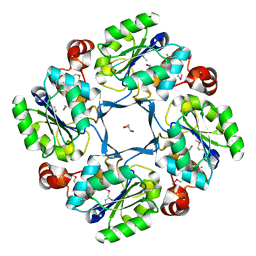
1J8D
 
 | |
1K1E
 
 | |
4GAH
 
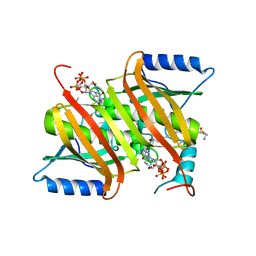 | | Human acyl-CoA thioesterases 4 in complex with undecan-2-one-CoA inhibitor | | Descriptor: | Thioesterase superfamily member 4, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3R)-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-[2-[(2R)-2-oxidanylundecyl]sulfanylethylamino]propyl]amino]butyl] hydrogen phosphate | | Authors: | Lim, K, Pathak, M.C, Herzberg, O. | | Deposit date: | 2012-07-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation of structure and function in the human hotdog-fold enzyme hTHEM4.
Biochemistry, 51, 2012
|
|
2HWG
 
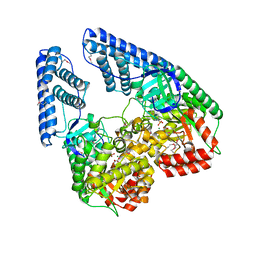 | | Structure of phosphorylated Enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system | | Descriptor: | MAGNESIUM ION, OXALATE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Lim, K, Teplyakov, A, Herzberg, O. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6XBZ
 
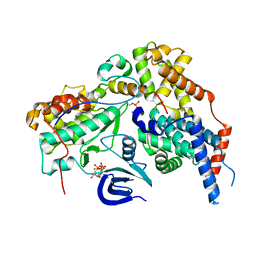 | | Structure of the human CDK-activating kinase | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Greber, B.J, Perez-Bertoldi, J.M, Lim, K, Iavarone, A.T, Toso, D.B, Nogales, E. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The cryoelectron microscopy structure of the human CDK-activating kinase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
