3J5P
 
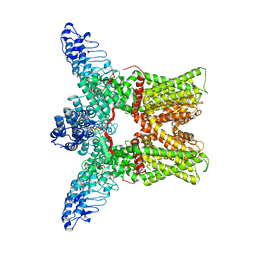 | |
3J5Q
 
 | | Structure of TRPV1 ion channel in complex with DkTx and RTX determined by single particle electron cryo-microscopy | | Descriptor: | Kappa-theraphotoxin-Cg1a 1, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Liao, M, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-04 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | TRPV1 structures in distinct conformations reveal activation mechanisms.
Nature, 504, 2013
|
|
3J5R
 
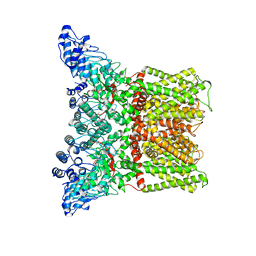 | |
7RIT
 
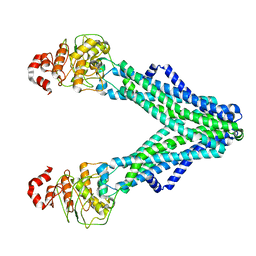 | | Drug-free A. baumannii MsbA | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-07-20 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7RSL
 
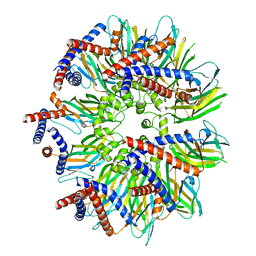 | | Seipin forms a flexible cage at lipid droplet formation sites | | Descriptor: | Seipin | | Authors: | Arlt, H, Sui, X, Folger, B, Adams, C, Chen, X, Remme, R, Hamprecht, F.A, DiMaio, F, Liao, M, Goodman, J.M, Farese Jr, R.V, Walther, T.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-02-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Seipin forms a flexible cage at lipid droplet formation sites.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TDT
 
 | |
8FL8
 
 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8FKJ
 
 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8F39
 
 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8F29
 
 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 2024
|
|
7MDX
 
 | | LolCDE nucleotide-free | | Descriptor: | (2R)-2-(tridecanoyloxy)propyl hexadecanoate, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | Authors: | Sharma, S, Liao, M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins
Nat Commun, 12, 2021
|
|
7R6N
 
 | | Exon-free state of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic trimeric construct | | Descriptor: | Group I intron, MAGNESIUM ION | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6M
 
 | | Post-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | Descriptor: | Group I intron, Ligated exon mimic of the Group I intron, MAGNESIUM ION | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6L
 
 | | 5 prime exon-free pre-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | Descriptor: | Group I intron, 3 prime fragment plus 3 prime exon, 5 prime fragment, ... | | Authors: | Thelot, F, Liu, D, Liao, M, Yin, P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
8ETM
 
 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(1S,4r)-4-{4-[(4S)-2-({[4-(trifluoromethoxy)phenyl]methyl}carbamoyl)imidazo[1,2-a]pyridin-6-yl]phenyl}cyclohexyl]acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8ESM
 
 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, {(1r,4r)-4-[4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl]cyclohexyl}acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
7MDY
 
 | | LolCDE nucleotide-bound | | Descriptor: | ADP ORTHOVANADATE, Lipo-releasing system transmembrane protein lolC, Lipoprotein transporter subunit LolE, ... | | Authors: | Sharma, S, Liao, M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-11 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins
Nat Commun, 12, 2021
|
|
7MET
 
 | | A. baumannii MsbA in complex with TBT1 decoupler | | Descriptor: | 2-(4-chlorobenzamido)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7MEW
 
 | | E. coli MsbA in complex with G247 | | Descriptor: | (2E)-3-{1-cyclopropyl-7-[(1S)-1-(3,6-dichloro-2-fluorophenyl)ethoxy]naphthalen-2-yl}prop-2-enoic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-10-06 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
6NJ8
 
 | | Encapsulin iron storage compartment from Quasibacillus thermotolerans | | Descriptor: | Encapsulating protein for a DyP-type peroxidase, targeting peptide | | Authors: | Orlando, B.J, Giessen, T.W, Chambers, M.G, Liao, M, Silver, P.A. | | Deposit date: | 2019-01-02 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Large protein organelles form a new iron sequestration system with high storage capacity.
Elife, 8, 2019
|
|
8ERC
 
 | |
6VYI
 
 | | Cryo-EM structure of human diacylglycerol O-acyltransferase 1 | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Sui, X, Wang, K, Gluchowski, N, Liao, M, Walther, C.T, Farese, V.R. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Nature, 581, 2020
|
|
6VXJ
 
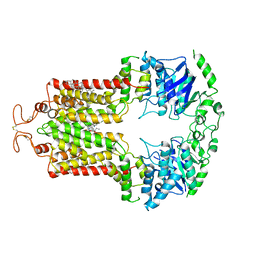 | | Structure of ABCG2 bound to SN38 | | Descriptor: | 7-ethyl-10-hydroxycamptothecin, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
6VXH
 
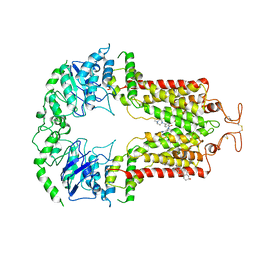 | | Structure of ABCG2 bound to imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
6VXI
 
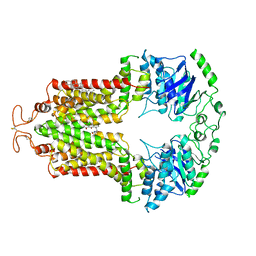 | | Structure of ABCG2 bound to mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
