1G3G
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Liao, H, Su, M, Yongkiettrakul, S, Byeon, I.-J.L, Tsai, M.-D. | | Deposit date: | 2000-10-24 | | Release date: | 2001-01-10 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structure of the FHA1 domain of yeast Rad53 and identification of binding sites for both FHA1 and its target protein Rad9
J.Mol.Biol., 304, 2000
|
|
1QU5
 
 | |
1DMZ
 
 | |
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
6KV2
 
 | |
1A8Y
 
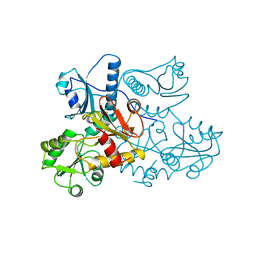 | | CRYSTAL STRUCTURE OF CALSEQUESTRIN FROM RABBIT SKELETAL MUSCLE SARCOPLASMIC RETICULUM AT 2.4 A RESOLUTION | | Descriptor: | CALSEQUESTRIN | | Authors: | Wang, S, Trumble, W.R, Liao, H, Wesson, C.R, Dunker, A.K, Kang, C. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum.
Nat.Struct.Biol., 5, 1998
|
|
1FHR
 
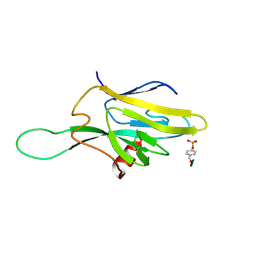 | | SOLUTION STRUCTURE OF THE FHA2 DOMAIN OF RAD53 COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Byeon, I.-J.L, Liao, H, Yongkiettrakul, S, Tsai, M.-D. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides.
J.Mol.Biol., 302, 2000
|
|
1FHQ
 
 | |
7YRN
 
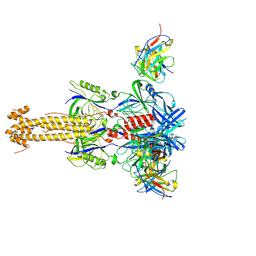 | |
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
4V6R
 
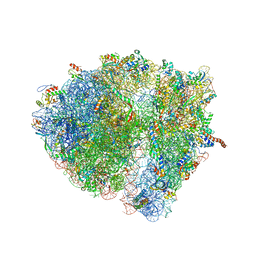 | | Structural characterization of mRNA-tRNA translocation intermediates (class 6 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6N
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (50S ribosome of class2 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6P
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 4b of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6S
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 3 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6O
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 4a of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7N28
 
 | |
7MXD
 
 | |
4V6Q
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 5 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1YQJ
 
 | | Crystal Structure of p38 Alpha in Complex with a Selective Pyridazine Inhibitor | | Descriptor: | 6((S)-3-BENZYLPIPERAZIN-1-YL)-3-(NAPHTHALEN-2-YL)-4-(PYRIDIN-4-YL)PYRAZINE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Tamayo, N, Liao, H, Goldberg, M, Syed, R, Li, V, Powers, D, Tudor, Y, Yu, V, Wong, M.L, Henkle, B, Middelton, S, Harvey, T, Jang, G, Hungate, R, Dominguez, C. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent pyridazine inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6WI5
 
 | |
4N0T
 
 | | Core structure of the U6 small nuclear ribonucleoprotein at 1.7 Angstrom resolution | | Descriptor: | SULFATE ION, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA | | Authors: | Montemayor, E.J, Curran, E.C, Liao, H, Andrews, K.L, Treba, C.N, Butcher, S.E, Brow, D.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Core structure of the U6 small nuclear ribonucleoprotein at 1.7- angstrom resolution.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3S2A
 
 | | Crystal structure of PI3K-gamma in complex with a quinoline inhibitor | | Descriptor: | N-{2-chloro-5-[4-(morpholin-4-yl)quinolin-6-yl]pyridin-3-yl}-4-fluorobenzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phospshoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors: Discovery and Structure-Activity Relationships of a Series of Quinoline and Quinoxaline Derivatives.
J.Med.Chem., 54, 2011
|
|
7E3J
 
 | |
3GZN
 
 | | Structure of NEDD8-activating enzyme in complex with NEDD8 and MLN4924 | | Descriptor: | NEDD8, NEDD8-activating enzyme E1 catalytic subunit, NEDD8-activating enzyme E1 regulatory subunit, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2009-04-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Mol.Cell, 37, 2010
|
|
