2XFL
 
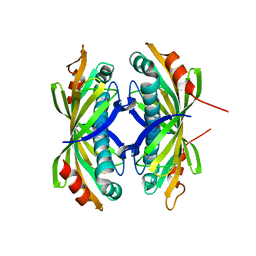 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Sun, H, Bricogne, G, Liang, Z, Lescar, J. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
1WT9
 
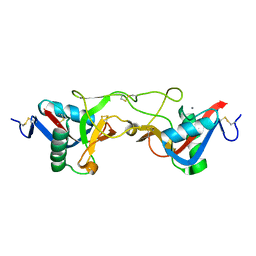 | | crystal structure of Aa-X-bp-I, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus | | Descriptor: | CALCIUM ION, agkisacutacin A chain, anticoagulant protein-B | | Authors: | Zhu, Z, Liu, S, Mo, X, Yu, X, Liang, Z, Zang, J, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2004-11-18 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterizations and Crystal structures of two snake venom proteins with the activity of binding coagulation factor X from Agkistrodon acutus
To be Published
|
|
1Y17
 
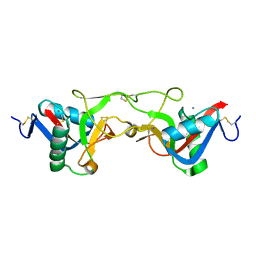 | | crystal structure of Aa-X-bp-II, a snake venom protein with the activity of binding to coagulation factor X from Agkistrodon acutus | | Descriptor: | CALCIUM ION, anticoagulant protein A, anticoagulant protein-B | | Authors: | Zhu, Z, Liu, S, Mo, X, Yu, X, Liang, Z, Zang, J, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2004-11-17 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterizations and Crystal structures of two snake venom proteins with the activity of binding coagulation factor X from Agkistrodon acutus
To be Published
|
|
4GXO
 
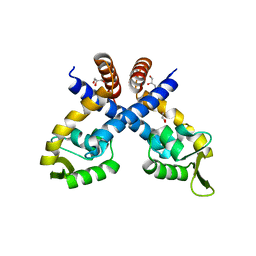 | | Crystal structure of Staphylococcus aureus protein SarZ mutant C13E | | Descriptor: | GLYCEROL, MarR family regulatory protein | | Authors: | Sun, F, Ding, Y, Ji, Q, Liang, Z, Deng, X, Wong, C.C, Yi, C, Zhang, L, Xie, S, Alvarez, S, Hicks, L.M, Luo, C, Jiang, H, Lan, L, He, C. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AMM
 
 | |
4AMP
 
 | |
4AMN
 
 | |
4AMO
 
 | |
1OP0
 
 | | Crystal Structure of AaV-SP-I, a Glycosylated Snake Venom Serine Proteinase from Agkistrodon acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Venom serine proteinase | | Authors: | Zhu, Z, Teng, M, Niu, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Amidolytic Activities of Two Glycosylated Snake Venom Serine Proteinases
J.BIOL.CHEM., 280, 2005
|
|
1OP2
 
 | | Crystal Structure of AaV-SP-II, a Glycosylated Snake Venom Serine Proteinase from Agkistrodon acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Venom serine proteinase | | Authors: | Zhu, Z, Teng, M, Niu, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Amidolytic Activities of Two Glycosylated Snake Venom Serine Proteinases
J.BIOL.CHEM., 280, 2005
|
|
2XEM
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | (3E,5E,7E,9E,11E,13E)-pentadeca-3,5,7,9,11,13-hexaen-2-one, DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Bricogne, G, Liang, Z.X, Lescar, J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
1W24
 
 | | Crystal Structure Of human Vps29 | | Descriptor: | VACUOLAR PROTEIN SORTING PROTEIN 29 | | Authors: | Wang, D, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2004-06-26 | | Release date: | 2005-03-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Vacuolar Protein Sorting Protein 29 Reveals a Phosphodiesterase/Nuclease-Like Fold and Two Protein-Protein Interaction Sites.
J.Biol.Chem., 280, 2005
|
|
7N44
 
 | | Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13) | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Reilly, R.A, Zhang, C.H, Deshmukh, M.G, Ippolito, J.A, Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency.
Acs Med.Chem.Lett., 12, 2021
|
|
8EMA
 
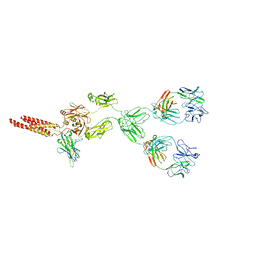 | | mouse full length B cell receptor | | Descriptor: | Anti-human Langerin 2G3 lambda chain, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Ying, D, Xiong, P, Michael, R. | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural principles of B cell antigen receptor assembly.
Nature, 612, 2022
|
|
8E4C
 
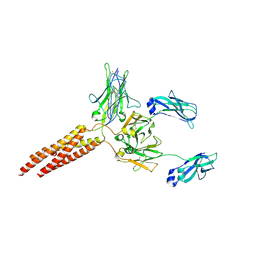 | | IgM BCR fab truncated form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain,Yellow fluorescent protein, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Dong, Y, Pi, X, Wu, H, Reth, M. | | Deposit date: | 2022-08-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural principles of B cell antigen receptor assembly.
Nature, 612, 2022
|
|
5IMX
 
 | |
8WUY
 
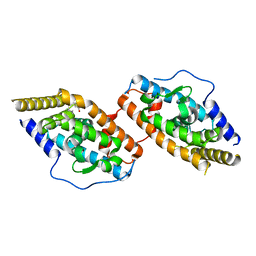 | | Crystal Structure of TR3 LBD in complex with para-positioned 3,4,5-trisubstituted benzene derivatives | | Descriptor: | Nuclear receptor subfamily 4immunitygroup A member 1, ~{N}-methyl-~{N}-octyl-3,4,5-tris(oxidanyl)benzamide | | Authors: | Hong, W.B, Chen, X.Q, Lin, T.W. | | Deposit date: | 2023-10-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and synthesis of anti-fibrotic compounds derived from para-positioned 3,4,5-trisubstituted benzene.
Bioorg.Chem., 144, 2024
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
7V61
 
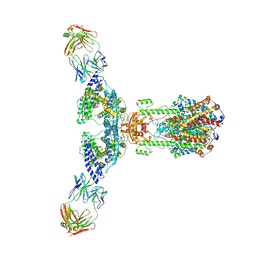 | | ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3E8, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker.
Signal Transduct Target Ther, 6, 2021
|
|
7L12
 
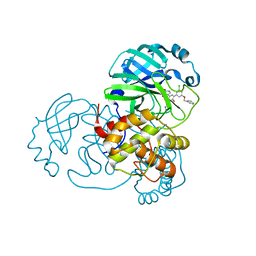 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 14 | | Descriptor: | (5S)-5-{3-[3-(benzyloxy)-5-chlorophenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L14
 
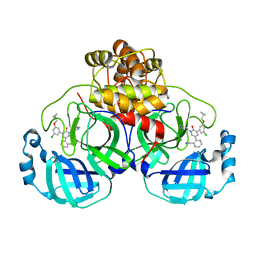 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 26 | | Descriptor: | 2-{3-[3-chloro-5-(cyclopropylmethoxy)phenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L13
 
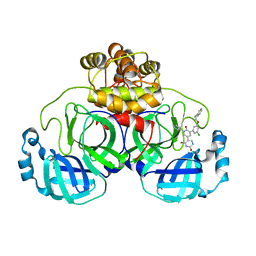 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5S)-5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]phenyl}-2-oxo[2H-[1,3'-bipyridine]]-5-yl)pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L10
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 (2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 2-[3-(3,5-dichlorophenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-13 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L11
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 2-[3-(3-chloro-5-propoxyphenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7CVA
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2 | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
