3BW3
 
 | | Crystal structures and site-directed mutagenesis study of nitroalkane oxidase from Streptomyces ansochromogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-nitropropane dioxygenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F. | | Deposit date: | 2008-01-08 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes
Biochem.Biophys.Res.Commun., 405, 2011
|
|
3BW2
 
 | | Crystal structures and site-directed mutagenesis study of nitroalkane oxidase from Streptomyces ansochromogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-nitropropane dioxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F. | | Deposit date: | 2008-01-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes
Biochem.Biophys.Res.Commun., 405, 2011
|
|
5D8F
 
 | |
5D8E
 
 | |
3BW4
 
 | |
8K80
 
 | |
8GPW
 
 | | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G | | Descriptor: | 1-[(~{Z})-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2~{S})-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxycyclopropane-1-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Song, D.Q, Li, Y.H. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G at 2.06 Angstroms resolution
To Be Published
|
|
6JBH
 
 | | Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter | | Descriptor: | TarG, TarH | | Authors: | Chen, L, Hou, W.T, Fan, T, Li, Y.H, Liu, B.H, Jiang, Y.L, Sun, L.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Mbio, 11, 2020
|
|
6JVN
 
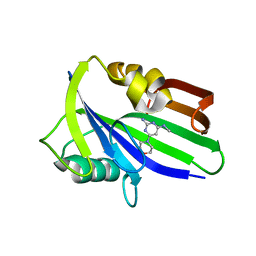 | | Crystal structure of human MTH1 in complex with compound MI1020 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-morpholin-4-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVS
 
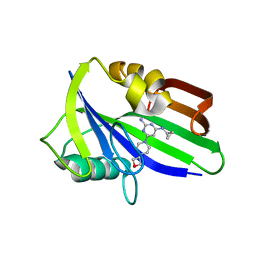 | | Crystal structure of human MTH1 in complex with compound MI1029 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-[4-(oxetan-3-yl)piperazin-1-yl]pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVK
 
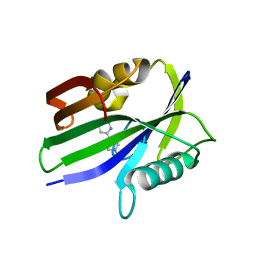 | | Crystal structure of human MTH1 in complex with compound MI1012 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVT
 
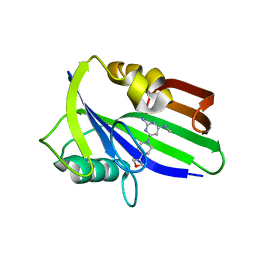 | | Crystal structure of human MTH1 in complex with compound MI1030 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-[4-(oxetan-3-yl)piperazin-1-yl]pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVJ
 
 | | Crystal structure of human MTH1 in complex with compound MI1006 | | Descriptor: | 5-ethyl-N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVR
 
 | | Crystal structure of human MTH1 in complex with compound MI1026 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVP
 
 | | Crystal structure of human MTH1 in complex with compound MI1024 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVQ
 
 | | Crystal structure of human MTH1 in complex with compound MI1025 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-morpholin-4-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVO
 
 | | Crystal structure of human MTH1 in complex with compound MI1022 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVL
 
 | | Crystal structure of human MTH1 in complex with compound MI1014 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-5-ethyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVM
 
 | | Crystal structure of human MTH1 in complex with compound MI1016 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-5-ethyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
7FH9
 
 | |
7FH4
 
 | | Chlorovirus PBCV-1 bi-functional dCMP/dCTP deaminase bi-DCD | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CMP/dCMP-type deaminase domain-containing protein, ... | | Authors: | She, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of a multi-functional deaminase in chlorovirus PBCV-1.
Arch.Biochem.Biophys., 727, 2022
|
|
5C2O
 
 | | Crystal structure of Streptococcus mutans Deoxycytidylate Deaminase complexed with dTTP | | Descriptor: | MAGNESIUM ION, Putative deoxycytidylate deaminase, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F, Dong, Y.H. | | Deposit date: | 2015-06-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of the allosteric regulation of Streptococcus mutans 2'-deoxycytidylate deaminase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7DLD
 
 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | Descriptor: | Carbonyl Reductase, MAGNESIUM ION | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLM
 
 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | Descriptor: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
