6L2W
 
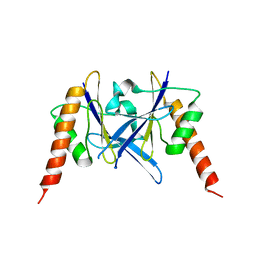 | | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1 | | Descriptor: | freshwater cyanophage protein | | Authors: | Wang, Y, Jin, H, Yang, F, Jiang, Y.L, Zhao, Y.Y, Chen, Z.P, Li, W.F, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-10-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1.
Proteins, 88, 2020
|
|
4PVD
 
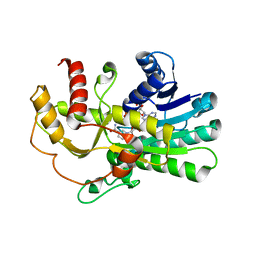 | | Crystal structure of yeast methylglyoxal/isovaleraldehyde reductase Gre2 complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent methylglyoxal reductase GRE2 | | Authors: | Guo, P.C, Bao, Z.Z, Li, W.F, Zhou, C.Z. | | Deposit date: | 2014-03-17 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast methylglyoxal/isovaleraldehyde reductase Gre2
Biochim.Biophys.Acta, 1844, 2014
|
|
6J3Q
 
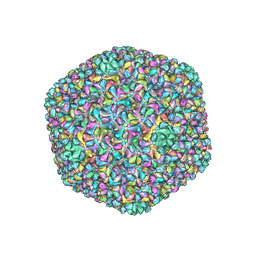 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | Descriptor: | cement protein, major capsid protein | | Authors: | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-05 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
6LRS
 
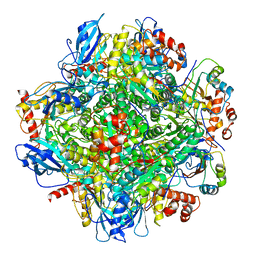 | | Cryo-EM structure of RbcL8-RbcS4 from Anabaena sp. PCC 7120 | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Sun, H, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-01-16 | | Release date: | 2020-07-15 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y2W
 
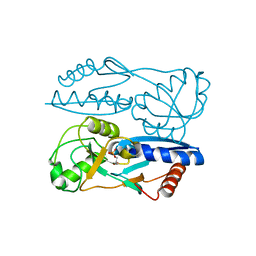 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
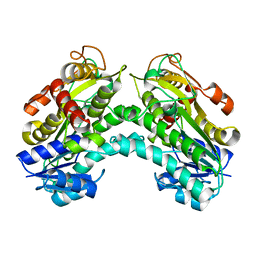
4PVC
 
 | |
4IJC
 
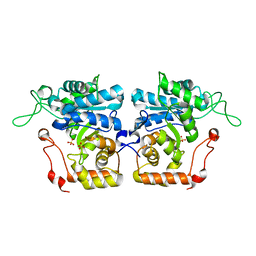 | | Crystal structure of arabinose dehydrogenase Ara1 from Saccharomyces cerevisiae | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, GLYCEROL, SULFATE ION | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-21 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IJR
 
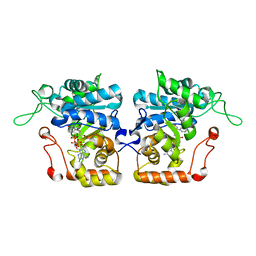 | | Crystal structure of Saccharomyces cerevisiae arabinose dehydrogenase Ara1 complexed with NADPH | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3CTF
 
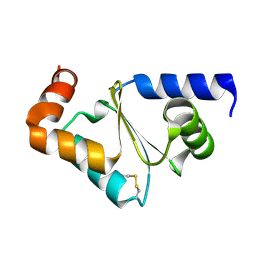 | | Crystal structure of oxidized GRX2 | | Descriptor: | Glutaredoxin-2 | | Authors: | Yu, J, Teng, Y.B, Zhou, C.Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the different activities of yeast Grx1 and Grx2.
Biochim.Biophys.Acta, 1804, 2010
|
|
3CTG
 
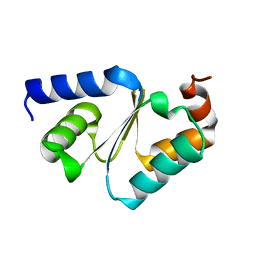 | |
8K3D
 
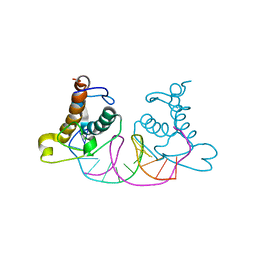 | | Crystal structure of NRF1 DBD bound to DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*CP*GP*CP*AP*TP*GP*CP*GP*CP*AP*CP*C)-3'), Nuclear respiratory factor 1 | | Authors: | Li, W.F, Liu, K, Min, J.R. | | Deposit date: | 2023-07-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of specific DNA sequence recognition by NRF1.
Nucleic Acids Res., 52, 2024
|
|
8HDS
 
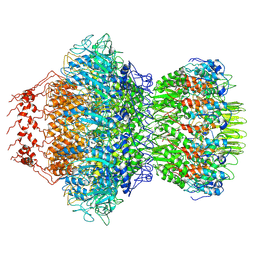 | | Cyanophage Pam3 portal-adaptor | | Descriptor: | Pam3 adaptor protein, Pam3 portal protein | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDT
 
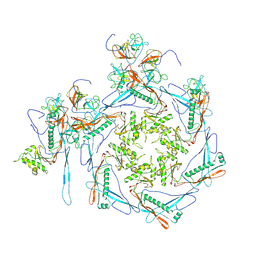 | |
8HDW
 
 | | Cyanophage Pam3 Sheath-tube | | Descriptor: | Pam3 sheath protein, pam3 tube | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDR
 
 | | Cyanophage Pam3 neck | | Descriptor: | Pam3 adaptor protein, Pam3 connector protein, Pam3 sheath protein, ... | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YHD
 
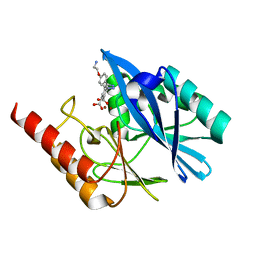 | | Crystal structure of VIM-2 MBL in complex with 3-(4-(4-(2-aminoethoxy)phenyl)-1H-1,2,3-triazol-1-yl)phthalic acid | | Descriptor: | 3-[4-[4-(2-azanylethoxy)phenyl]-1,2,3-triazol-1-yl]phthalic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YH9
 
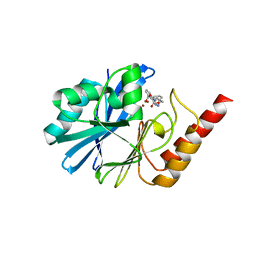 | | Crystal structure of IMP-1 MBL in complex with 3-(4-benzyl-1H-1,2,3-triazol-1-yl)phthalic acid | | Descriptor: | 3-[4-(phenylmethyl)-1,2,3-triazol-1-yl]phthalic acid, Beta-lactamase class B IMP-1, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YHC
 
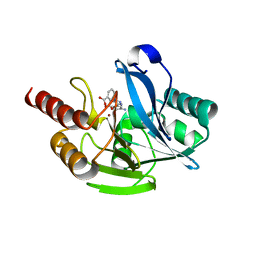 | | Crystal structure of VIM-2 MBL in complex with 3-(4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl)phthalic acid | | Descriptor: | 3-[4-(3-aminophenyl)-1,2,3-triazol-1-yl]phthalic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YHA
 
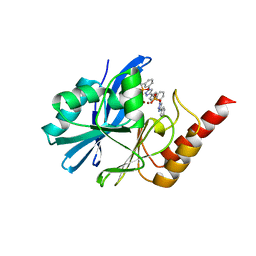 | | Crystal structure of IMP-1 MBL in complex with (3-(4-(p-tolyl)-1H-1,2,3-triazol-1-yl)benzyl)phosphonic acid | | Descriptor: | Beta-lactamase class B IMP-1, ZINC ION, [3-[4-(4-methylphenyl)-1,2,3-triazol-1-yl]phenyl]methylphosphonic acid | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YHB
 
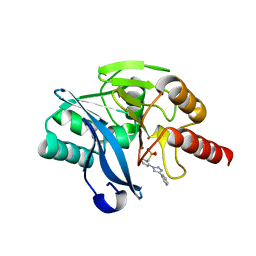 | | Crystal structure of VIM-2 MBL in complex with (2-(4-phenyl-1H-1,2,3-triazol-1-yl)benzyl)phosphonic acid | | Descriptor: | Beta-lactamase class B VIM-2, ZINC ION, [2-(4-phenyl-1,2,3-triazol-1-yl)phenyl]methylphosphonic acid | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7EEQ
 
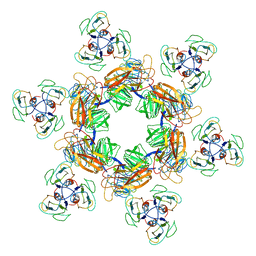 | | Cyanophage Pam1 tail machine | | Descriptor: | Needle head proteins, Tailspike head-binding domain | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEL
 
 | | Cyanophage Pam1 capsid asymmetric unit | | Descriptor: | Cement (decoration) proteins, Major capsid proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEP
 
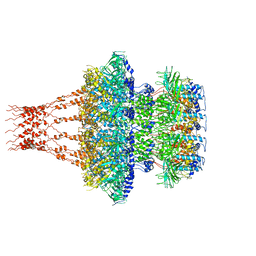 | | Cyanophage Pam1 portal-adaptor complex | | Descriptor: | Pam1 adaptor proteins, Pam1 portal proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEA
 
 | |
