1AKH
 
 | | MAT A1/ALPHA2/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MATING-TYPE PROTEIN A-1), ... | | Authors: | Li, T, Jin, Y, Vershon, A.K, Wolberger, C. | | Deposit date: | 1997-05-19 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract.
Nucleic Acids Res., 26, 1998
|
|
1KU5
 
 | | Crystal Structure of recombinant histone HPhA from hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, HPhA, SULFATE ION | | Authors: | Li, T, Sun, F, Ji, X, Feng, Y, Rao, Z. | | Deposit date: | 2002-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii
J.MOL.BIOL., 325, 2003
|
|
1YRN
 
 | | CRYSTAL STRUCTURE OF THE MATA1/MATALPHA2 HOMEODOMAIN HETERODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*TP*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*TP*AP*TP*TP*AP*C P*AP*TP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*AP*TP*GP*TP*AP*AP*TP*AP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT A1 HOMEODOMAIN), ... | | Authors: | Li, T, Stark, M.R, Johnson, A.D, Wolberger, C. | | Deposit date: | 1995-11-02 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA.
Science, 270, 1995
|
|
7VPG
 
 | | Crystal structure of the C-terminal tail of SARS-CoV-1 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | Descriptor: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | Authors: | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | Deposit date: | 2021-10-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
7VPH
 
 | | Crystal structure of the C-terminal tail of SARS-CoV-2 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | Descriptor: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | Authors: | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | Deposit date: | 2021-10-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
6DTP
 
 | |
1MYM
 
 | |
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
2B5L
 
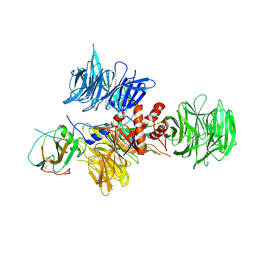 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | Descriptor: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2B5N
 
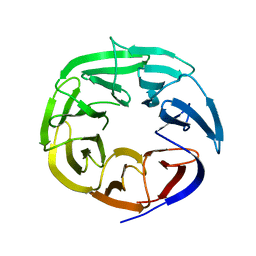 | | Crystal Structure of the DDB1 BPB Domain | | Descriptor: | ISOPROPYL ALCOHOL, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
2B5M
 
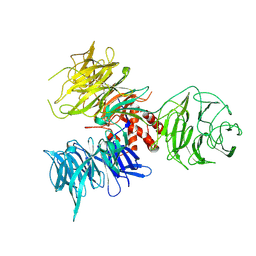 | | Crystal Structure of DDB1 | | Descriptor: | damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
3I7P
 
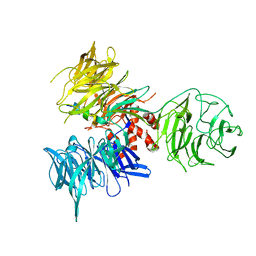 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR40A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 40A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I7O
 
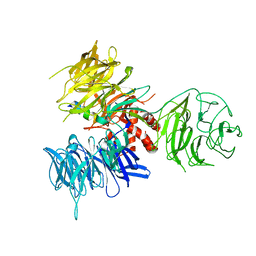 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of IQWD1 | | Descriptor: | DNA damage-binding protein 1, IQ motif and WD repeat-containing protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I7H
 
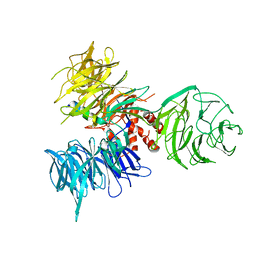 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of HBX | | Descriptor: | DNA damage-binding protein 1, X protein | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I7N
 
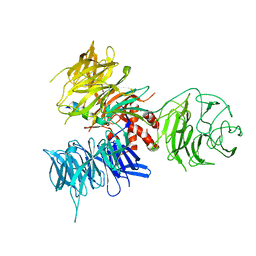 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDTC1 | | Descriptor: | DNA damage-binding protein 1, WD and tetratricopeptide repeats protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I7L
 
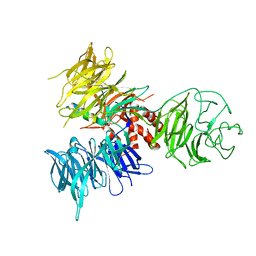 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of DDB2 | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I7K
 
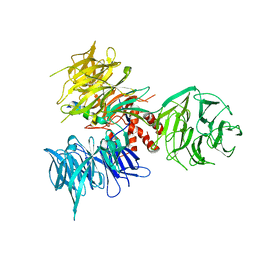 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WHX | | Descriptor: | DNA damage-binding protein 1, X protein | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I8E
 
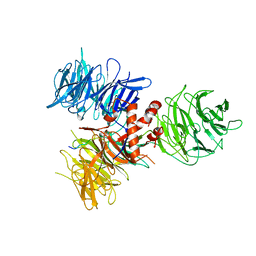 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR42A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 42A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I89
 
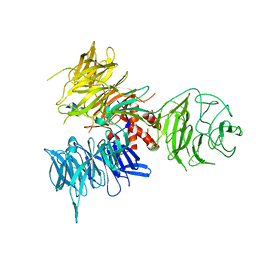 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR22 | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 22 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I8C
 
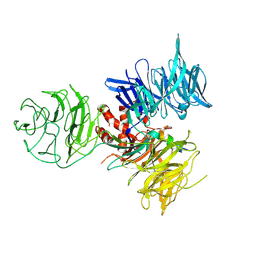 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR21A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 21A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8IAG
 
 | |
6JAU
 
 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
