4L22
 
 | |
2G0J
 
 | | Crystal structure of SMU.848 from Streptococcus mutans | | Descriptor: | hypothetical protein SMU.848 | | Authors: | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
2G0I
 
 | | Crystal structure of SMU.848 from Streptococcus mutans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, hypothetical protein SMU.848 | | Authors: | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-08-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
3CXP
 
 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 mutant E156A | | Descriptor: | CHLORIDE ION, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3CXQ
 
 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3CXS
 
 | | Crystal structure of human GNA1 | | Descriptor: | Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3S70
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO solved by As-SAD | | Descriptor: | ACETATE ION, CACODYLATE ION, Caspase-6, ... | | Authors: | Su, X.-D, Liu, X, Wang, X.-J. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Get phases from arsenic anomalous scattering: de novo SAD phasing of two protein structures crystallized in cacodylate buffer
Plos One, 6, 2011
|
|
4IYR
 
 | | Crystal structure of full-length caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.-J, Li, L.-F, Su, X.-D. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BJV
 
 | |
3NR2
 
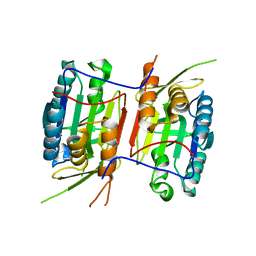 | | Crystal structure of Caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Su, X.-D, Wang, X.-J, Liu, X, Mi, W, Wang, K.-T. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
3OD5
 
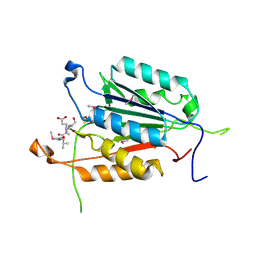 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO | | Descriptor: | CACODYLATE ION, Caspase-6, peptide aldehyde inhibitor AC-VEID-CHO | | Authors: | Wang, X.-J, Liu, X, Wang, K.-T, Cao, Q, Su, X.-D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
3OIX
 
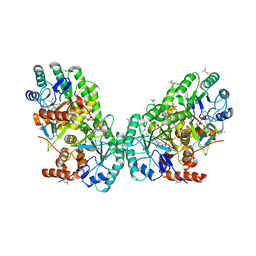 | | Crystal structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Putative dihydroorotate dehydrogenase; dihydroorotate oxidase | | Authors: | Liu, Y, Gao, Z.Q, Liu, C.P, Dong, Y.H. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans
Acta Crystallogr.,Sect.F, 67, 2011
|
|
