1LBG
 
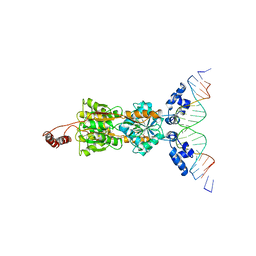 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBH
 
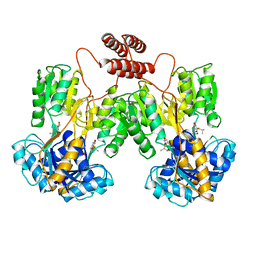 | | INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBI
 
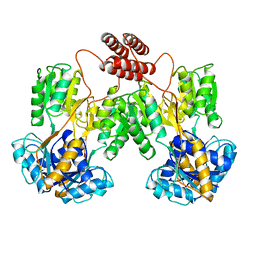 | | LAC REPRESSOR | | Descriptor: | LAC REPRESSOR | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
2FJR
 
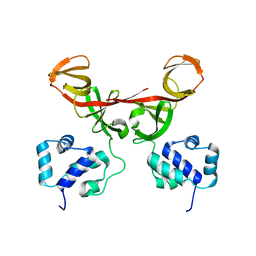 | | Crystal Structure of Bacteriophage 186 | | Descriptor: | Repressor protein CI | | Authors: | Lewis, M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of cooperative regulation at an alternate genetic switch
Mol.Cell, 21, 2006
|
|
2FKD
 
 | |
1AFS
 
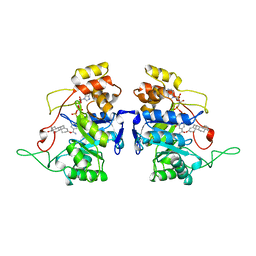 | | RECOMBINANT RAT LIVER 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE (3-ALPHA-HSD) COMPLEXED WITH NADP AND TESTOSTERONE | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TESTOSTERONE | | Authors: | Bennett, M.J, Albert, R.H, Jez, J.M, Ma, H, Penning, T.M, Lewis, M. | | Deposit date: | 1997-03-13 | | Release date: | 1997-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
Structure, 5, 1997
|
|
1LRP
 
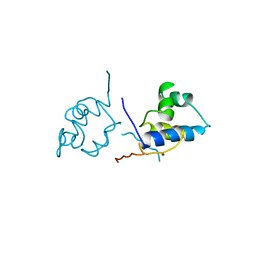 | |
1EFA
 
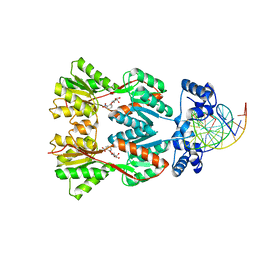 | |
1RAL
 
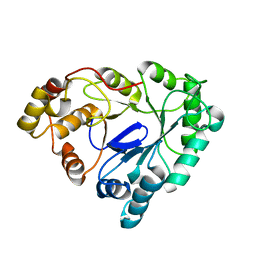 | | THREE-DIMENSIONAL STRUCTURE OF RAT LIVER 3ALPHA-HYDROXYSTEROID(SLASH)DIHYDRODIOL DEHYDROGENASE: A MEMBER OF THE ALDO-KETO REDUCTASE SUPERFAMILY | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Hoog, S.S, Pawlowski, J.E, Alzari, P.M, Penning, T.M, Lewis, M. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase: a member of the aldo-keto reductase superfamily.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1LWI
 
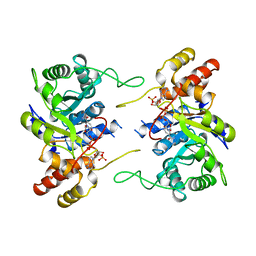 | | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE FROM RATTUS NORVEGICUS | | Descriptor: | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M.J, Schlegel, B.P, Jez, J.M, Penning, T.M, Lewis, M. | | Deposit date: | 1996-02-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
Biochemistry, 35, 1996
|
|
3BDN
 
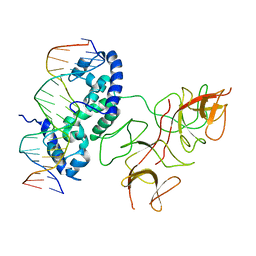 | | Crystal Structure of the Lambda Repressor | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DAP*DCP*DCP*DAP*DCP*DTP*DGP*DGP*DCP*DGP*DGP*DTP*DGP*DAP*DTP*DAP*DT)-3'), DNA (5'-D(*DTP*DAP*DTP*DAP*DTP*DCP*DAP*DCP*DCP*DGP*DCP*DCP*DAP*DGP*DTP*DGP*DGP*DTP*DAP*DT)-3'), Lambda Repressor | | Authors: | Stayrook, S.E, Jaru-Ampornpan, P, Hochschild, A, Lewis, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.909 Å) | | Cite: | Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding
Nature, 452, 2008
|
|
1F39
 
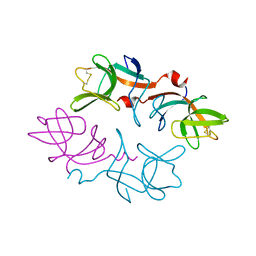 | | CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR C-TERMINAL DOMAIN | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Bell, C.E, Frescura, P, Hochschild, A, Lewis, M. | | Deposit date: | 2000-06-01 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding.
Cell(Cambridge,Mass.), 101, 2000
|
|
2PE5
 
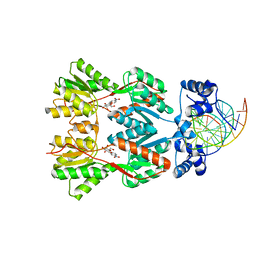 | | Crystal Structure of the Lac Repressor bound to ONPG in repressed state | | Descriptor: | 2-nitrophenyl beta-D-galactopyranoside, DNA (5'-D(*DAP*DAP*DTP*DTP*DGP*DTP*DGP*DAP*DGP*DCP*DGP*DCP*DTP*DCP*DAP*DCP*DAP*DAP*DTP*DT)-3'), Lactose operon repressor | | Authors: | Daber, R, Stayrook, S.E, Rosenberg, A, Lewis, M. | | Deposit date: | 2007-04-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of lac repressor bound to allosteric effectors
J.Mol.Biol., 370, 2007
|
|
1IHI
 
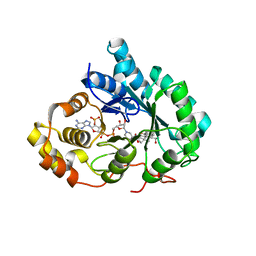 | | Crystal Structure of Human Type III 3-alpha-Hydroxysteroid Dehydrogenase/Bile Acid Binding Protein (AKR1C2) Complexed with NADP+ and Ursodeoxycholate | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, ISO-URSODEOXYCHOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jin, Y, Stayrook, S.E, Albert, R.H, Palackal, N.T, Penning, T.M, Lewis, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human type III 3alpha-hydroxysteroid dehydrogenase/bile acid binding protein complexed with NADP(+) and ursodeoxycholate.
Biochemistry, 40, 2001
|
|
1JYE
 
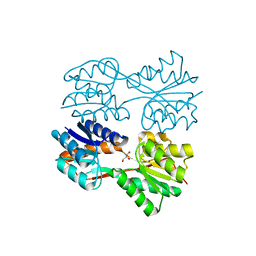 | | Structure of a Dimeric Lac Repressor with C-terminal Deletion and K84L Substitution | | Descriptor: | GLYCEROL, Lactose Operon Repressor | | Authors: | Bell, C.E, Barry, J, Matthews, K.S, Lewis, M. | | Deposit date: | 2001-09-12 | | Release date: | 2001-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a variant of lac repressor with increased thermostability and decreased affinity for operator.
J.Mol.Biol., 313, 2001
|
|
1JWL
 
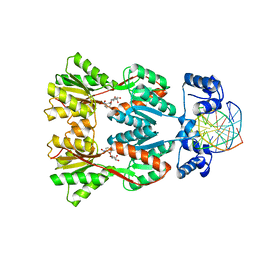 | | Structure of the Dimeric lac Repressor/Operator O1/ONPF Complex | | Descriptor: | 2-nitrophenyl beta-D-fucopyranoside, 5'-D(*AP*GP*AP*AP*T*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*T)-3', 5'-D(*TP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3', ... | | Authors: | Bell, C.E, Lewis, M. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystallographic analysis of Lac repressor bound to natural operator O1.
J.Mol.Biol., 312, 2001
|
|
1JYF
 
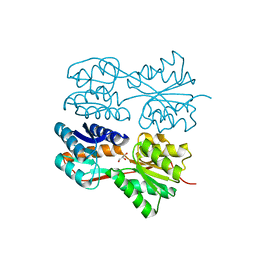 | | Structure of the Dimeric Lac Repressor with an 11-residue C-terminal Deletion. | | Descriptor: | GLYCEROL, Lactose Operon Repressor | | Authors: | Bell, C.E, Barry, J, Matthews, K.S, Lewis, M. | | Deposit date: | 2001-09-12 | | Release date: | 2001-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a variant of lac repressor with increased thermostability and decreased affinity for operator.
J.Mol.Biol., 313, 2001
|
|
1KCA
 
 | |
1M3W
 
 | | Crystal Structure of a Molecular Maquette Scaffold | | Descriptor: | H10H24, MERCURY (II) ION | | Authors: | Huang, S.S, Gibney, B.R, Stayrook, S.E, Dutton, P.L, Lewis, M. | | Deposit date: | 2002-07-01 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Structure of a Maquette Scaffold
J.Mol.Biol., 326, 2003
|
|
2YNO
 
 | | yeast betaprime COP 1-304H6 | | Descriptor: | COATOMER SUBUNIT BETA', POLY ALA | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2YNP
 
 | | yeast betaprime COP 1-604 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.962 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2YNN
 
 | | yeast betaprime COP 1-304 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF, SULFATE ION | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
4BV4
 
 | | Structure and allostery in Toll-Spatzle recognition | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN SPAETZLE C-106, PROTEIN TOLL, ... | | Authors: | Lewis, M.F, Arnot, C.J, Beeston, H, McCoy, A, Ashcroft, A.E, Gay, N.J, Gangloff, M. | | Deposit date: | 2013-06-24 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cytokine Spatzle Binds to the Drosophila Immunoreceptor Toll with a Neurotrophin-Like Specificity and Couples Receptor Activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1AAQ
 
 | |
3MO7
 
 | |
