1D8A
 
 | | E. COLI ENOYL REDUCTASE/NAD+/TRICLOSAN COMPLEX | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Levy, C.W, Roujeinikova, A, Sedelnikova, S, Baker, P.J, Stuitje, A.R, Slabas, A.R, Rice, D.W, Rafferty, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 1999-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of triclosan activity.
Nature, 398, 1999
|
|
1KKR
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE CONTAINING (2S,3S)-3-METHYLASPARTIC ACID | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, 3-METHYLASPARTATE AMMONIA-LYASE, MAGNESIUM ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, K, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
1KKO
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
8QHE
 
 | | Crystal structure IR-09 | | Descriptor: | 6-phosphogluconate dehydrogenase NADP-binding domain-containing protein, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biocatalysis in Drug Design: Engineered Reductive Aminases (RedAms) Are Used to Access Chiral Building Blocks with Multiple Stereocenters.
J.Am.Chem.Soc., 145, 2023
|
|
6QIX
 
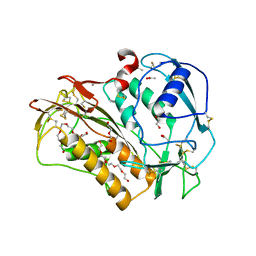 | | The crystal structure of Trichuris muris p43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The major secreted protein of the whipworm parasite tethers to matrix and inhibits interleukin-13 function.
Nat Commun, 10, 2019
|
|
5O4L
 
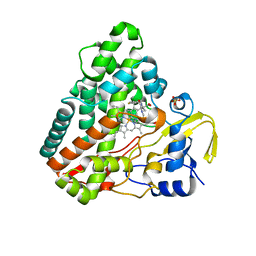 | | Crystal structure of P450 CYP121 in complex with compound 6a. | | Descriptor: | 1-[(4-fluorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-05-29 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
5O4K
 
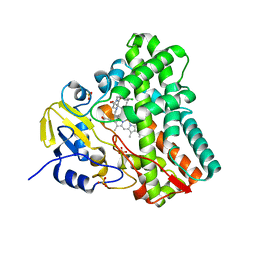 | | Crystal structure of P450 CYP121 in complex with compound 6b. | | Descriptor: | 1-[(4-chlorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-05-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
6I3C
 
 | |
6I3D
 
 | |
5OP9
 
 | | The crystal structure of P450 CYP121 in complex with lead compound 7e | | Descriptor: | 4-(imidazol-1-ylmethyl)-3-(4-methoxyphenyl)-1-phenyl-pyrazole, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
5OPA
 
 | | The crystal structure of P450 CYP121 in complex with lead compound 7b | | Descriptor: | 3-(4-fluorophenyl)-4-(imidazol-1-ylmethyl)-1-phenyl-pyrazole, DI(HYDROXYETHYL)ETHER, Mycocyclosin synthase, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
8BOU
 
 | | Crystal structure of Blautia producta GH94 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Blautia producta is a competent degrader among human gut Firmicutes for utilizing dietary beta mixed linkage glucan
To Be Published
|
|
7ZRN
 
 | |
5CPO
 
 | | Crystal structure of XenA from Pseudomonas putida in complex with an NADH mimic (mBu) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 1-butyl-1,4,5,6-tetrahydropyridine-3-carboxamide, CALCIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
6QB2
 
 | |
6RNW
 
 | |
6RNV
 
 | |
6FOZ
 
 | |
6FOY
 
 | |
6FP1
 
 | | The crystal structure of P.fluorescens Kynurenine 3-monooxygenase (KMO) in complex with competitive inhibitor No. 1 | | Descriptor: | 2-(6-chloranyl-5,7-dimethyl-3-oxidanylidene-1,4-benzoxazin-4-yl)ethanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Levy, C.W, Leys, D. | | Deposit date: | 2018-02-08 | | Release date: | 2019-08-21 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites.
Commun Biol, 2, 2019
|
|
6FPH
 
 | | The crystal structure of P.fluorescens Kynurenine 3-monooxygenase (KMO) in complex with competitive inhibitor No. 1h | | Descriptor: | 6-chloranyl-5,7-dimethyl-4-(1~{H}-1,2,3,4-tetrazol-5-ylmethyl)-1,4-benzoxazin-3-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Levy, C.W, Leys, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-21 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A brain-permeable inhibitor of the neurodegenerative disease target kynurenine 3-monooxygenase prevents accumulation of neurotoxic metabolites.
Commun Biol, 2, 2019
|
|
6FP0
 
 | |
6FOX
 
 | |
6GEO
 
 | |
6GEQ
 
 | |
