4I5E
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase in complex with NADP+ | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5D
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase in its apo form | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase, SULFATE ION | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5F
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5G
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S, A86N, S88A | | Descriptor: | Alclohol dehydrogenase/short-chain dehydrogenase | | Authors: | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | Deposit date: | 2012-11-28 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4GRX
 
 | | Structure of an omega-aminotransferase from Paracoccus denitrificans | | Descriptor: | Aminotransferase, DELTA-AMINO VALERIC ACID, SODIUM ION | | Authors: | Rausch, C, Lerchner, A, Schiefner, A, Skerra, A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the omega-aminotransferase from Paracoccus denitrificans and its phylogenetic relationship with other class III aminotransferases that have biotechnological potential.
Proteins, 81, 2013
|
|
3K5C
 
 | | Human BACE-1 complex with NB-216 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Macrocyclic BACE-1 inhibitors acutely reduce Abeta in brain after po application.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8SBJ
 
 | | Co-structure Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform complexed with brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(1H-pyrazol-3-yl)-3H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
8SBC
 
 | | Co-structure of Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform and brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(pyridin-2-yl)-1H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
6FYK
 
 | | X-Ray structure of CLK2-KD(136-496)/Indazole1 at 2.39A | | Descriptor: | (2~{S})-4-methyl-1-[5-(3-methyl-2~{H}-indazol-5-yl)pyridin-3-yl]oxy-pentan-2-amine, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYL
 
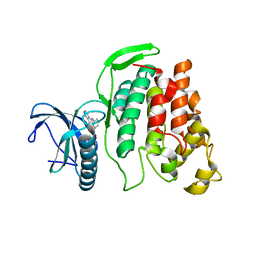 | | X-ray structure of CLK2-KD(136-496)/CX-4945 at 1.95A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYV
 
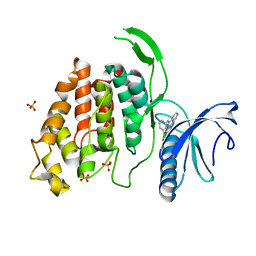 | | X-RAY STRUCTURE OF CLK4-KD(146-480)/CX-4945 AT 2.46A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK4, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYO
 
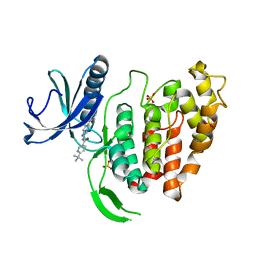 | | X-RAY STRUCTURE OF CLK1-KD(148-484)/Cpd-2 AT 2.32A | | Descriptor: | 6-~{tert}-butyl-~{N}-[6-(1~{H}-pyrazol-4-yl)-1~{H}-imidazo[1,2-a]pyridin-2-yl]pyridine-3-carboxamide, Dual specificity protein kinase CLK1, SULFATE ION | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYP
 
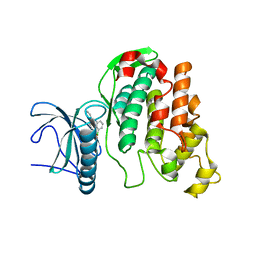 | | X-RAY STRUCTURE OF CLK3-KD(GP-[275-632], NON-PHOS.)/CX-4945 AT 2.29A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYR
 
 | | X-RAY STRUCTURE OF CLK3-KD(GP-[275-632], NON-PHOS.)/Cpd-2 AT 1.42A | | Descriptor: | 6-~{tert}-butyl-~{N}-[6-(1~{H}-pyrazol-4-yl)-1~{H}-imidazo[1,2-a]pyridin-2-yl]pyridine-3-carboxamide, Dual specificity protein kinase CLK3 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FYI
 
 | | X-ray Structure of CLK2-KD(130-496)/TG003 at 2.6A | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
3QBH
 
 | | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors | | Descriptor: | (4S)-4-(2-hydroxy-5-{[(3S,4S,5R)-4-hydroxy-1,1-dioxido-5-{[3-(propan-2-yl)benzyl]amino}tetrahydro-2H-thiopyran-3-yl]methyl}benzyl)-3-propyl-1,3-oxazolidin-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5QD7
 
 | | Crystal structure of BACE complex with BMC014 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatric yclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
