3QR3
 
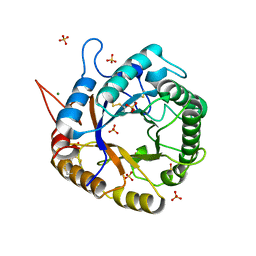 | | Crystal Structure of Cel5A (EG2) from Hypocrea jecorina (Trichoderma reesei) | | Descriptor: | Endoglucanase EG-II, MAGNESIUM ION, SULFATE ION | | Authors: | Lee, T.M, Farrow, M.F, Kaiser, J.T, Arnold, F.H, Mayo, S.L. | | Deposit date: | 2011-02-16 | | Release date: | 2011-11-02 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural study of Hypocrea jecorina Cel5A.
Protein Sci., 20, 2011
|
|
2RIF
 
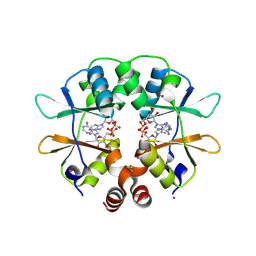 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CESIUM ION, Conserved protein with 2 CBS domains | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon.
J.Mol.Biol., 380, 2008
|
|
2RIH
 
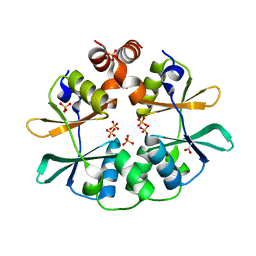 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, Conserved protein with 2 CBS domains, SULFATE ION | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon
J.Mol.Biol., 380, 2008
|
|
3NYZ
 
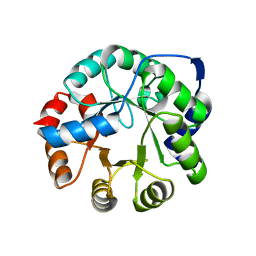 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 | | Descriptor: | Indole-3-glycerol phosphate synthase, SULFATE ION | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NYD
 
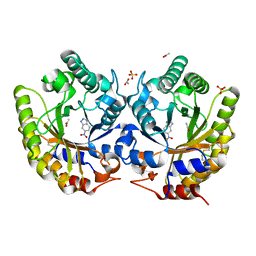 | | Crystal Structure of Kemp Eliminase HG-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-14 | | Release date: | 2011-06-29 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NZ1
 
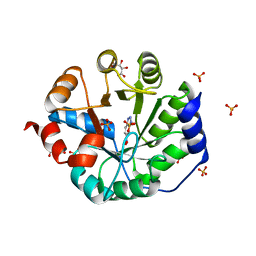 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, Indole-3-glycerol phosphate synthase, L(+)-TARTARIC ACID, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XFT
 
 | |
3O2L
 
 | |
