4MAC
 
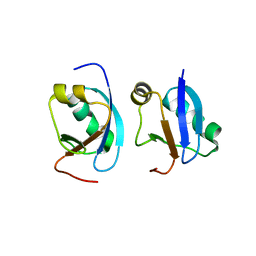 | | Crystal structure of CIDE-N domain of FSP27 | | Descriptor: | Cell death activator CIDE-3 | | Authors: | Park, H.H, Lee, S.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for homo-dimerization of the CIDE domain revealed by the crystal structure of the CIDE-N domain of FSP27
Biochem.Biophys.Res.Commun., 439, 2013
|
|
1IZ3
 
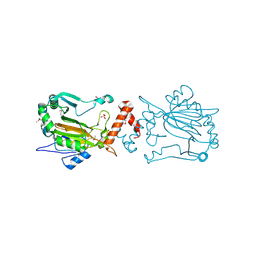 | | Dimeric structure of FIH (Factor inhibiting HIF) | | Descriptor: | FIH, SULFATE ION | | Authors: | Lee, C, Kim, S.-J, Jeong, D.-G, Lee, S.M, Ryu, S.-E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau.
J.Biol.Chem., 278, 2003
|
|
5FPZ
 
 | | The structure of KdgF from Yersinia enterocolitica with malonate bound in the active site. | | Descriptor: | MALONIC ACID, NICKEL (II) ION, PECTIN DEGRADATION PROTEIN | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FPX
 
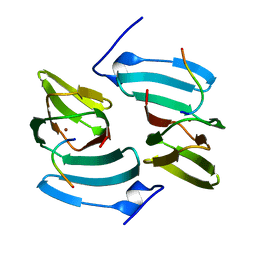 | | The structure of KdgF from Yersinia enterocolitica. | | Descriptor: | NICKEL (II) ION, PECTIN DEGRADATION PROTEIN, PEPTIDE | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FQ0
 
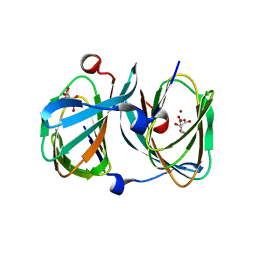 | | The structure of KdgF from Halomonas sp. | | Descriptor: | CITRATE ANION, KDGF, NICKEL (II) ION, ... | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3KTU
 
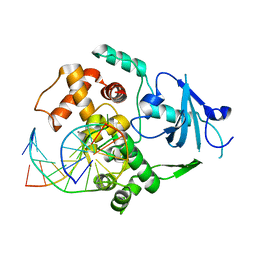 | |
6PGQ
 
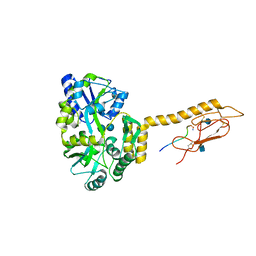 | |
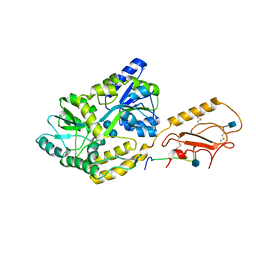
6PFO
 
 | |
6L2K
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_R49E | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), SULFATE ION | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L2Z
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pnuemoniae_D191G | | Descriptor: | AMMONIUM ION, GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-07 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L2I
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_WT | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L2S
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_D83G | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)) | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L2R
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_E195S | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)) | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
7UVS
 
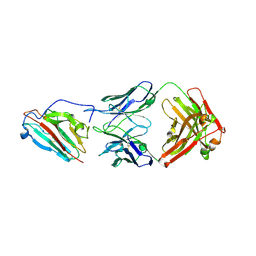 | | Pfs230 domain 1 bound by LMIV230-02 Fab | | Descriptor: | Gametocyte surface protein P230, LMIV230-02 Fab heavy chain, LMIV230-02 Fab light chain, ... | | Authors: | Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UVH
 
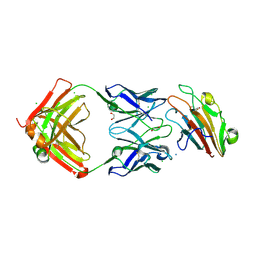 | | Pfs230 domain 1 bound by RUPA-32 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMMONIUM ION, ... | | Authors: | Ivanochko, D, Newton, J, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UVO
 
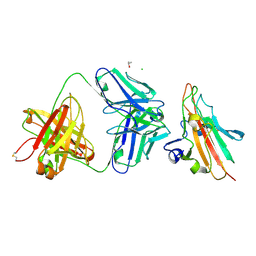 | | Pfs230 domain 1 bound by RUPA-38 Fab | | Descriptor: | CHLORIDE ION, Gametocyte surface protein P230, ISOPROPYL ALCOHOL, ... | | Authors: | Ivanochko, D, Newton, J, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UVQ
 
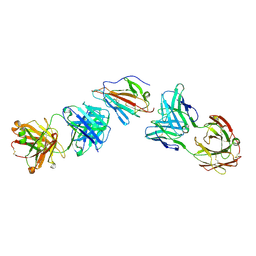 | | Pfs230 domain 1 bound by RUPA-97 and 15C5 Fabs | | Descriptor: | Fab Heavy Chain, Fab Kappa Light Chain, Gametocyte surface protein P230 | | Authors: | Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
7UVI
 
 | | Pfs230 domain 1 bound by RUPA-55 Fab | | Descriptor: | Gametocyte surface protein P230, RUPA-55 Fab heavy chain, RUPA-55 Fab light chain | | Authors: | Ivanochko, D, Newton, J, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
5YN3
 
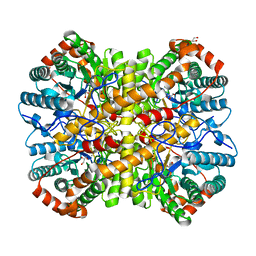 | |
5Z6T
 
 | |
5Z6U
 
 | |
6D1U
 
 | | Crystal structure of the human CLR:RAMP1 extracellular domain heterodimer in complex with adrenomedullin 2/intermedin | | Descriptor: | ADM2, Maltose-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, SODIUM ION, ... | | Authors: | Pioszak, A, Roehrkasse, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analyses reveal a triple beta-turn receptor-bound conformation of adrenomedullin 2/intermedin and enable peptide antagonist design.
J. Biol. Chem., 293, 2018
|
|
