5Y7K
 
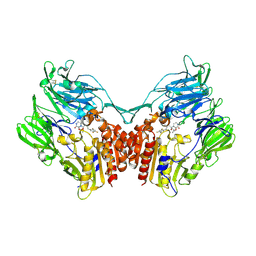 | | Crystal structure of human DPP4 in complex with inhibitor1 | | Descriptor: | (R)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazine-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7J
 
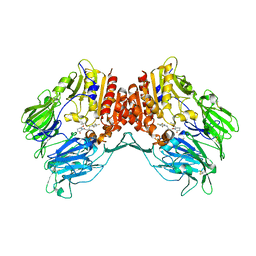 | | Crystal structure of human DPP4 in complex with inhibitor2 | | Descriptor: | (S)-4-((R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl)-3-(tert-butoxymethyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
5Y7H
 
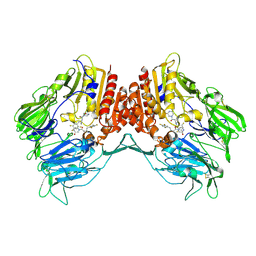 | | Crystal structure of human DPP4 in complex with inhibitor3 | | Descriptor: | (R)-4-(3-amino-4-(2,4,5-trifluorophenyl)butanoyl)piperazin-2-one, Dipeptidyl peptidase 4 | | Authors: | Lee, H.K, Kim, E.E. | | Deposit date: | 2017-08-17 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unique binding mode of Evogliptin with human dipeptidyl peptidase IV.
Biochem.Biophys.Res.Commun., 494, 2017
|
|
2OS1
 
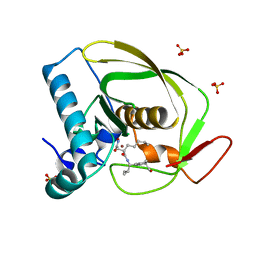 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
2OS3
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | ACTINONIN, COBALT (II) ION, Peptide deformylase | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Parh, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
2OS0
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
8F4O
 
 | | Apo structure of the TPP riboswitch aptamer domain | | Descriptor: | IRIDIUM HEXAMMINE ION, TETRAETHYLENE GLYCOL, TPP riboswitch aptamer domain, ... | | Authors: | Lee, H.-K, Wang, Y.-X, Stagno, J.R. | | Deposit date: | 2022-11-11 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state.
Structure, 31, 2023
|
|
5GPG
 
 | | Co-crystal structure of the FK506 binding domain of human FKBP25, Rapamycin and the FRB domain of human mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP3, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Lee, H.B, Lee, S.Y, Rhee, H.W, Lee, C.W. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Acs Cent.Sci., 2, 2016
|
|
7D0Z
 
 | |
7D0X
 
 | |
7D0Y
 
 | |
7E4E
 
 | |
7VM9
 
 | |
7YF7
 
 | |
6M6K
 
 | |
6M0C
 
 | |
6M0B
 
 | |
6M6J
 
 | |
6MSY
 
 | | Anti-HIV-1 Fab Fab 2G12 + Man4 re-refinement | | Descriptor: | ACETATE ION, Fab 2G12, light chain, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6MUB
 
 | | Anti-HIV-1 Fab 2G12 + Man5 re-refinement | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Wilson, I.A, Calarese, D.A, Stanfield, R.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6MNF
 
 | | Anti-HIV-1 Fab 2G12 + Man8 re-refinement | | Descriptor: | Fab 2G12, light chain, Fab 2g12, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6MU3
 
 | | Anti-HIV-1 Fab 2G12 + Man7 re-refinement | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Wilson, I.A, Calarese, D.A, Stanfield, R.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
