1IE6
 
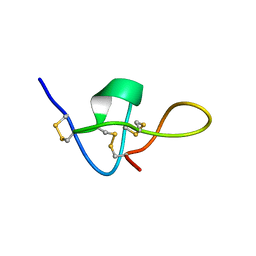 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | Descriptor: | IMPERATOXIN A | | Authors: | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | Deposit date: | 2001-04-07 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
5H1Z
 
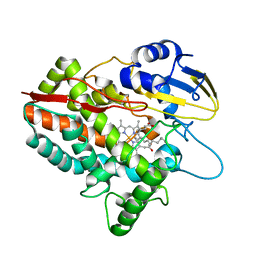 | | CYP153D17 from Sphingomonas sp. PAMC 26605 | | Descriptor: | DODECANE, PROTOPORPHYRIN IX CONTAINING FE, putative CYP alkane hydroxylase CYP153D17 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Putative Cytochrome P450 Alkane Hydroxylase (CYP153D17) from Sphingomonas sp. PAMC 26605 and Its Conformational Substrate Binding
Int J Mol Sci, 17, 2016
|
|
5IT1
 
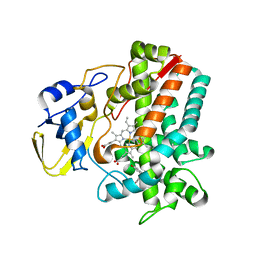 | | Streptomyces peucetius CYP105P2 complex with biphenyl compound | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-03-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450 (CYP105P2) from Streptomyces peucetius and Its Conformational Changes in Response to Substrate Binding
Int J Mol Sci, 17, 2016
|
|
6A4E
 
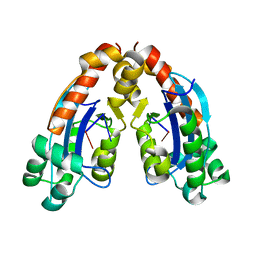 | |
6A4D
 
 | |
6A4A
 
 | |
6A4F
 
 | |
1T1T
 
 | | Solution Structure of Kurtoxin | | Descriptor: | Kurtoxin | | Authors: | Lee, C.W, Min, H.J, Cho, E.M, Kohno, T, Eu, Y.J, Kim, J.I. | | Deposit date: | 2004-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kurtoxin
To be Published
|
|
1LA4
 
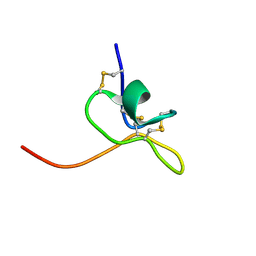 | | Solution Structure of SGTx1 | | Descriptor: | SGTx1 | | Authors: | Lee, C.W, Roh, S.H, Kim, S, Endoh, H, Kodera, Y, Maeda, T, Swartz, K.J, Kim, J.I. | | Deposit date: | 2002-03-28 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Characterization of SGTx1, a Modifier of Kv2.1 Channel Gating
Biochemistry, 43, 2004
|
|
6JZL
 
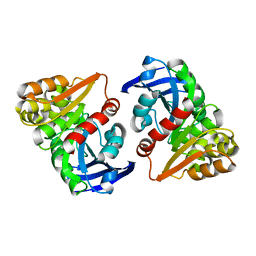 | |
7C4X
 
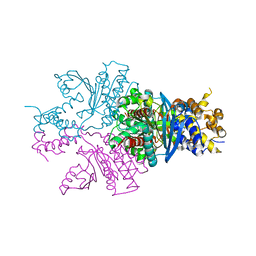 | |
7DVN
 
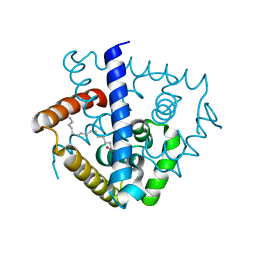 | | Crystal structure of a MarR family protein in complex with a lipid-like effector molecule from the psychrophilic bacterium Paenisporosarcina sp. TG-14 | | Descriptor: | MarR family transcriptional regulator, PALMITIC ACID | | Authors: | Lee, C.W, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a MarR family protein from the psychrophilic bacterium Paenisporosarcina sp. TG-14 in complex with a lipid-like molecule.
Iucrj, 8, 2021
|
|
5WQ0
 
 | | Receiver domain of Spo0A from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Stage 0 sporulation protein | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
J. Microbiol., 55, 2017
|
|
5YX4
 
 | | Isoliquiritigenin-complexed Chalcone isomerase (S189A) from the Antarctic vascular plant Deschampsia Antarctica (DaCHI1) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Chalcone-flavonone isomerase family protein | | Authors: | Lee, C.W, Park, S, Lee, J.H. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzymatic properties of chalcone isomerase from the Antarctic vascular plant Deschampsia antarctica Desv.
PLoS ONE, 13, 2018
|
|
5YX3
 
 | |
5BVT
 
 | | Palmitate-bound pFABP5 | | Descriptor: | Epidermal fatty acid-binding protein, PALMITOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVS
 
 | | Linoleate-bound pFABP4 | | Descriptor: | Fatty acid-binding protein, LINOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVQ
 
 | | Ligand-unbound pFABP4 | | Descriptor: | fatty acid-binding protein | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5Z2E
 
 | |
5Z2F
 
 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
5Z2D
 
 | |
5H3H
 
 | | Esterase (EaEST) from Exiguobacterium antarcticum | | Descriptor: | Abhydrolase domain-containing protein, ETHANEPEROXOIC ACID | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-11 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum.
Plos One, 12, 2017
|
|
5XNT
 
 | | Structure of CYP106A2 from Bacillus sp. PAMC 23377 | | Descriptor: | Cytochrome P450 CYP106, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Kim, K.-H, Bikash, D, Park, S.-H, Park, H, Oh, T.-J, Lee, J.H. | | Deposit date: | 2017-05-24 | | Release date: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of a Cytochrome P450 (BaCYP106A2) fromBacillussp. PAMC 23377.
J. Microbiol. Biotechnol., 27, 2017
|
|
6A7J
 
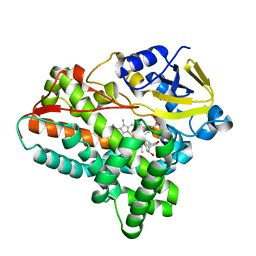 | | Testerone bound CYP154C4 from Streptomyces sp. ATCC 11861 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, TESTOSTERONE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
6A7I
 
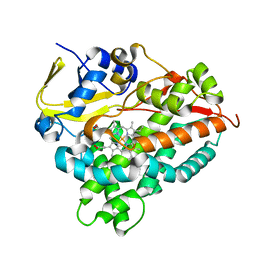 | | CYP154C4 from Streptomyces sp. W2061 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
