5U4K
 
 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | Descriptor: | CREB-binding protein, Transcription factor p65 | | Authors: | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
5URN
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the transactivation domain 1 of p65 | | Descriptor: | RNA polymerase II transcription factor B subunit 1, Transcription factor p65 | | Authors: | Lecoq, L, Omichinski, J.G, Raiola, L, Cyr, N, Chabot, P, Arseneault, G, Legault, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-03-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
3ZG4
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase | | Descriptor: | ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Dubee, V, Triboulet, S, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZGP
 
 | | NMR structure of the catalytic domain from E. faecium L,D- transpeptidase acylated by ertapenem | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ERFK/YBIS/YCFS/YNHG | | Authors: | Lecoq, L, Triboulet, S, Dubee, V, Bougault, C, Hugonnet, J.E, Arthur, M, Simorre, J.P. | | Deposit date: | 2012-12-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Structure of Enterococcus Faecium L,D---Transpeptidase Acylated by Ertapenem Provides Insight Into the Inactivation Mechanism.
Acs Chem.Biol., 8, 2013
|
|
3ZQD
 
 | | B. subtilis L,D-transpeptidase | | Descriptor: | L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J.-P, Bougault, C, Arthur, M, Hugonnet, J.-E, Veckerle, C, Pessey, O. | | Deposit date: | 2011-06-09 | | Release date: | 2012-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
4A52
 
 | | NMR structure of the imipenem-acylated L,D-transpeptidase from Bacillus subtilis | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, PUTATIVE L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J, Bougault, C, Arthur, M, Hugonnet, J, Veckerle, C, Pessey, O. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-30 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
2N23
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the N-terminal activation domain of EKLF (TAD1) | | Descriptor: | Krueppel-like factor 1, RNA polymerase II transcription factor B subunit 1 | | Authors: | Lecoq, L, Morse, T, Raiola, L, Arseneault, G, Omichinski, J. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Complex between the N-terminal Transactivation Domain of EKLF and the p62/Tfb1 subunit of TFIIH
To be Published
|
|
5C17
 
 | | Crystal structure of the mercury-bound form of MerB2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-13 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0U
 
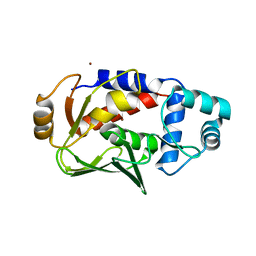 | | Crystal structure of the copper-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, COPPER (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0T
 
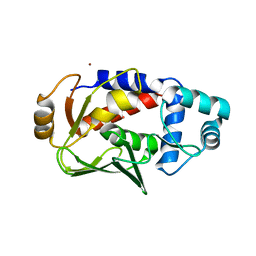 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5DSF
 
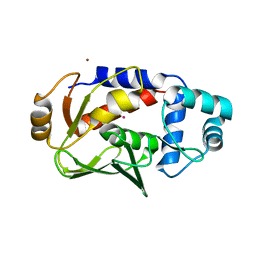 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
2N0Y
 
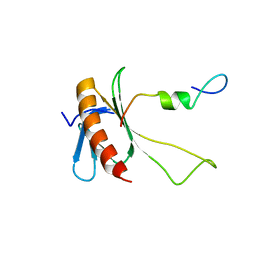 | | NMR structure of the complex between the C-terminal domain of the Rift Valley fever virus protein NSs and the PH domain of the Tfb1 subunit of TFIIH | | Descriptor: | Non-structural protein NS-S, RNA polymerase II transcription factor B subunit 1 | | Authors: | Cyr, N, de la Fuente, C, Lecoq, L, Guendel, I, Chabot, P.R, Kehn-Hall, K, Omichinski, J.G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Omega XaV motif in the Rift Valley fever virus NSs protein is essential for degrading p62, forming nuclear filaments and virulence.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8C0O
 
 | |
8AAC
 
 | |
