4DUR
 
 | | The X-ray Crystal Structure of Full-Length type II Human Plasminogen | | Descriptor: | ACETATE ION, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Law, R.H.P, Caradoc-Davies, T, Whisstock, J.C. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The X-ray crystal structure of full-length human plasminogen
Cell Rep, 1, 2012
|
|
4DUU
 
 | |
5UGG
 
 | | Protease Inhibitor | | Descriptor: | Nalpha-[trans-4-(aminomethyl)cyclohexane-1-carbonyl]-N-octyl-O-[(quinolin-2-yl)methyl]-L-tyrosinamide, Plasminogen | | Authors: | Law, R.H.P, Wu, G, Whisstock, J.C. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-31 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray crystal structure of plasmin with tranexamic acid-derived active site inhibitors.
Blood Adv, 1, 2017
|
|
5UGD
 
 | | Protease Inhibitor | | Descriptor: | Nalpha-[trans-4-(aminomethyl)cyclohexane-1-carbonyl]-N-octyl-O-[(pyridin-4-yl)methyl]-L-tyrosinamide, Plasminogen | | Authors: | Law, R.H.P, Wu, G, Whisstock, J.C. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | X-ray crystal structure of plasmin with tranexamic acid-derived active site inhibitors.
Blood Adv, 1, 2017
|
|
2R9Y
 
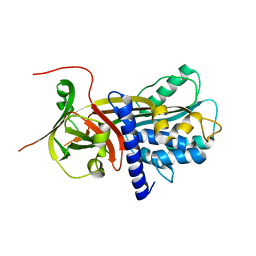 | | Structure of antiplasmin | | Descriptor: | Alpha-2-antiplasmin | | Authors: | Law, R.H.P, Sofian, T, Kan, W.T, Horvath, A.J, Hitchen, C.R, Langendorf, C.G, Buckle, A.M, Whisstock, J.C, Coughlin, P.B. | | Deposit date: | 2007-09-14 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray crystal structure of the fibrinolysis inhibitor {alpha}2-antiplasmin
Blood, 111, 2008
|
|
6DN5
 
 | |
6DN8
 
 | | SPRY domain-containing SOCS box protein 2 complexed with (GZJ)VDINNN(CY3) Cyclic peptide inhibitor | | Descriptor: | (GZJ)VDINNN(CY3) Cyclic peptide inhibitor, ACETATE ION, SPRY domain-containing SOCS box protein 4, ... | | Authors: | Law, R.H.P, Caradoc-Davies, T.T, Norton, R.S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-04-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Cyclic Peptide Inhibitor of the iNOS-SPSB Protein-Protein Interaction as a Potential Anti-Infective Agent.
ACS Chem. Biol., 13, 2018
|
|
5UG6
 
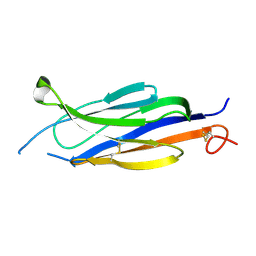 | | Perforin C2 Domain - T431D | | Descriptor: | IODIDE ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
5UG7
 
 | | Calcium bound Perforin C2 Domain - T431D | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
6NMB
 
 | |
6OG4
 
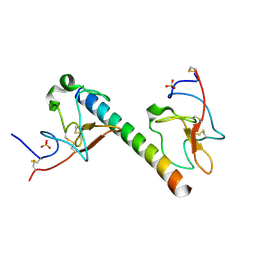 | | plasminogen binding group A streptococcal M protein | | Descriptor: | Plasminogen, Plasminogen-binding group A streptococcal M-like protein PAM, SULFATE ION | | Authors: | Law, R.H.P, Quek, A.J, Whisstock, J.C, Caradoc-Davies, T.T. | | Deposit date: | 2019-04-01 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function Characterization of the a1a2 Motifs of Streptococcus pyogenes M Protein in Human Plasminogen Binding.
J.Mol.Biol., 431, 2019
|
|
6CXO
 
 | | Complement component-9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement component C9, ... | | Authors: | Law, R.H.P, Spicer, B.A, Caradoc-Davies, T.T. | | Deposit date: | 2018-04-03 | | Release date: | 2018-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The first transmembrane region of complement component-9 acts as a brake on its self-assembly.
Nat Commun, 9, 2018
|
|
6D3Z
 
 | | Protease SFTI complex | | Descriptor: | Plasminogen, Trypsin inhibitor 1 | | Authors: | Law, R.H.P, Wu, G. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Highly Potent and Selective Plasmin Inhibitors Based on the Sunflower Trypsin Inhibitor-1 Scaffold Attenuate Fibrinolysis in Plasma.
J. Med. Chem., 62, 2019
|
|
6DN7
 
 | |
6D40
 
 | |
6D3Y
 
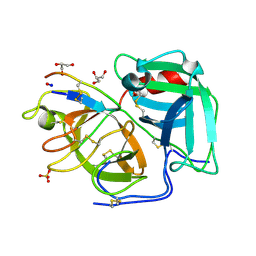 | |
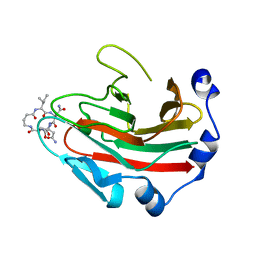
6DN6
 
 | |
6D3X
 
 | |
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3A
 
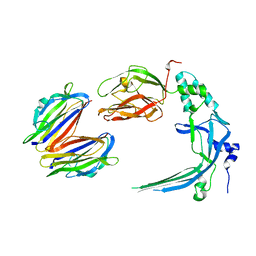 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3M
 
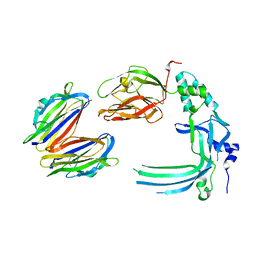 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3N
 
 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
5FMW
 
 | | The poly-C9 component of the Complement Membrane Attack Complex | | Descriptor: | POLYC9 | | Authors: | Dudkina, N.V, Spicer, B.A, Reboul, C.F, Conroy, P.J, Lukoyanova, N, Elmlund, H, Law, R.H.P, Ekkel, S.M, Kondos, S.C, Goode, R.J.A, Ramm, G, Whisstock, J.C, Saibil, H.R, Dunstone, M.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-17 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the Poly-C9 Component of the Complement Membrane Attack Complex
Nat.Commun., 7, 2016
|
|
3VP6
 
 | | Structural characterization of Glutamic Acid Decarboxylase; insights into the mechanism of autoinactivation | | Descriptor: | 4-oxo-4H-pyran-2,6-dicarboxylic acid, GLYCEROL, Glutamate decarboxylase 1 | | Authors: | Langendorf, C.G, Tuck, K.L, Key, T.L.G, Rosado, C.J, Wong, A.S.M, Fenalti, G, Buckle, A.M, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates
Biosci.Rep., 33, 2013
|
|
3ZLH
 
 | | Structure of group A Streptococcal enolase | | Descriptor: | ENOLASE | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
