7ACN
 
 | | CRYSTAL STRUCTURES OF ACONITASE WITH ISOCITRATE AND NITROISOCITRATE BOUND | | Descriptor: | ACONITASE, IRON/SULFUR CLUSTER, ISOCITRIC ACID | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1991-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of aconitase with isocitrate and nitroisocitrate bound.
Biochemistry, 31, 1992
|
|
1GXS
 
 | | Crystal Structure of Hydroxynitrile Lyase from Sorghum bicolor in Complex with Inhibitor Benzoic Acid: a novel cyanogenic enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZOIC ACID, DECANOIC ACID, ... | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Wajant, H, Effenberger, F. | | Deposit date: | 2002-04-11 | | Release date: | 2002-10-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Hydroxynitrile Lyase from Sorghum Bicolor in Complex with the Inhibitor Benzoic Acid: A Novel Cyanogenic Enzyme
Biochemistry, 41, 2002
|
|
1NIS
 
 | | CRYSTAL STRUCTURE OF ACONITASE WITH TRANS-ACONITATE AND NITROCITRATE BOUND | | Descriptor: | 2-HYDROXY-2-NITROMETHYL SUCCINIC ACID, ACONITASE, IRON/SULFUR CLUSTER | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1993-01-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of aconitase with trans-aconitate and nitrocitrate bound.
J.Mol.Biol., 237, 1994
|
|
1NIT
 
 | | CRYSTAL STRUCTURE OF ACONITASE WITH TRANS-ACONITATE AND NITROCITRATE BOUND | | Descriptor: | ACONITASE, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1993-01-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of aconitase with trans-aconitate and nitrocitrate bound.
J.Mol.Biol., 237, 1994
|
|
8ACN
 
 | | CRYSTAL STRUCTURES OF ACONITASE WITH ISOCITRATE AND NITROISOCITRATE BOUND | | Descriptor: | ACONITASE, IRON/SULFUR CLUSTER, NITROISOCITRIC ACID | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1991-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of aconitase with isocitrate and nitroisocitrate bound.
Biochemistry, 31, 1992
|
|
1E89
 
 | | ON THE MECHANISM OF CYANOGENESIS CATALYZED BY HYDROXYNITRILE LYASE FROM MANIHOT ESCULENTA. CRYSTAL STRUCTURE OF ACTIVE SITE MUTANT SER80ALA IN COMPLEX WITH ACETONE CYANOHYDRIN | | Descriptor: | HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Wajant, H, Effenberger, F. | | Deposit date: | 2000-09-18 | | Release date: | 2001-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic Aspects of Cyanogenesis from Active-Site Mutant Ser80Ala of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Acetone Cyanohydrin
Protein Sci., 10, 2001
|
|
1EB9
 
 | | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot esculenta | | Descriptor: | HYDROXYNITRILE LYASE, P-HYDROXYBENZALDEHYDE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Kobler, C, Wajant, H, Effenberger, F. | | Deposit date: | 2001-07-24 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot Esculenta.
Protein Sci., 11, 2002
|
|
1EB8
 
 | | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot esculenta | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-ACETONE-CYANOHYDRIN LYASE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Kobler, C, Wajant, H, Effenberger, F. | | Deposit date: | 2001-07-24 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Determinants of Substrate Specificity of Hydroxynitrile Lyase from Manihot Esculenta.
Protein Sci., 11, 2002
|
|
1E8D
 
 | | MECHANISTIC ASPECTS OF CYANOGENESIS FROM ACTIVE SITE MUTANT SER80ALA OF HYDROXYNITRILE LYASE FROM MANIHOT ESCULENTA IN COMPLEX WITH ACETONE CYANOHYDRIN | | Descriptor: | 2-HYDROXY-2-METHYLPROPANENITRILE, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Wajant, H, Effenberger, F. | | Deposit date: | 2000-09-19 | | Release date: | 2001-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Aspects of Cyanogenesis from Active-Site Mutant Ser80Ala of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Acetone Cyanohydrin.
Protein Sci., 10, 2001
|
|
1DWO
 
 | | Crystal Structure of Hydroxynitrile Lyase from Manihot esculenta in Complex with Substrates Acetone and Chloroacetone:Implications for the Mechanism of Cyanogenesis | | Descriptor: | ACETONE, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Forster, S, Miehlich, B, Wajant, H, Effenberger, F. | | Deposit date: | 1999-12-10 | | Release date: | 2000-12-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Substrates Acetone and Chloroacetone: Implications for the Mechanism of Cyanogenesis
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DWQ
 
 | | Crystal Structure of Hydroxynitrile Lyase from Manihot esculenta in Complex with Substrates Acetone and Chloroacetone:Implications for the Mechanism of Cyanogenesis | | Descriptor: | CHLOROACETONE, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Forster, S, Mielich, B, Wajant, H, Effenberger, F. | | Deposit date: | 1999-12-10 | | Release date: | 2000-12-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Substrates Acetone and Chloroacetone: Implications for the Mechanism of Cyanogenesis
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DWP
 
 | | Crystal Structure of Hydroxynitrile Lyase from Manihot esculenta at 2.2 Angstrom Resolution | | Descriptor: | ACETATE ION, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Wagner, U, Kratky, C, Mielich, B, Wajant, H, Forster, S, Effenberger, F. | | Deposit date: | 1999-12-10 | | Release date: | 2000-12-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Substrates Acetone and Chloroacetone: Implications for the Mechanism of Cyanogenesis
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FGH
 
 | | COMPLEX WITH 4-HYDROXY-TRANS-ACONITATE | | Descriptor: | 4-HYDROXY-ACONITATE ION, ACONITASE, IRON/SULFUR CLUSTER | | Authors: | Lauble, H, Kennedy, M.C, Emptage, M.H, Beinert, H, Stout, C.D. | | Deposit date: | 1996-09-14 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The reaction of fluorocitrate with aconitase and the crystal structure of the enzyme-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3ANA
 
 | |
1B0M
 
 | | ACONITASE R644Q:FLUOROCITRATE COMPLEX | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, PROTEIN (ACONITASE) | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-11 | | Release date: | 1998-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1B0J
 
 | | CRYSTAL STRUCTURE OF ACONITASE WITH ISOCITRATE | | Descriptor: | ACONITATE HYDRATASE, IRON/SULFUR CLUSTER, ISOCITRIC ACID | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-10 | | Release date: | 1998-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex
Protein Sci., 8, 1999
|
|
1B0K
 
 | | S642A:FLUOROCITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, OXYGEN ATOM, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1C96
 
 | | S642A:CITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1C97
 
 | | S642A:ISOCITRATE COMPLEX OF ACONITASE | | Descriptor: | IRON/SULFUR CLUSTER, ISOCITRIC ACID, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1AMI
 
 | |
1AMJ
 
 | |
1ACO
 
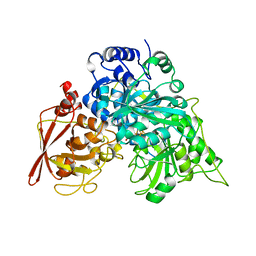 | | CRYSTAL STRUCTURE OF ACONITASE WITH TRANSACONITATE BOUND | | Descriptor: | ACONITASE, ACONITATE ION, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D. | | Deposit date: | 1993-01-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of aconitase with trans-aconitate and nitrocitrate bound.
J.Mol.Biol., 237, 1994
|
|
9DNA
 
 | |
2D25
 
 | | C-C-A-G-G-C-M5C-T-G-G; HELICAL FINE STRUCTURE, HYDRATION, AND COMPARISON WITH C-C-A-G-G-C-C-T-G-G | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*(5CM)P*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Heinemann, U, Hahn, M. | | Deposit date: | 1991-04-23 | | Release date: | 1991-04-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G.
J.Biol.Chem., 267, 1992
|
|
1D26
 
 | |
