1A7S
 
 | | ATOMIC RESOLUTION STRUCTURE OF HBP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Karlsen, S, Iversen, L.F, Larsen, I.K, Flodgaard, H.J, Kastrup, J.S. | | Deposit date: | 1998-03-17 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic resolution structure of human HBP/CAP37/azurocidin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AE5
 
 | | HUMAN HEPARIN BINDING PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEPARIN BINDING PROTEIN | | Authors: | Iversen, L.F, Kastrup, J.S, Bjorn, S.E, Rasmussen, P.B, Wiberg, F.C, Flodgaard, H.J, Larsen, I.K. | | Deposit date: | 1997-03-05 | | Release date: | 1998-03-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HBP, a multifunctional protein with a serine proteinase fold.
Nat.Struct.Biol., 4, 1997
|
|
1QZ1
 
 | | Crystal Structure of the Ig 1-2-3 fragment of NCAM | | Descriptor: | Neural cell adhesion molecule 1, 140 kDa isoform | | Authors: | Soroka, V, Kolkova, K, Kastrup, J.S, Diederichs, K, Breed, J, Kiselyov, V.V, Poulsen, F.M, Larsen, I.K, Welte, W, Berezin, V, Bock, E, Kasper, C. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion
Structure, 11, 2003
|
|
1FY1
 
 | | [R23S,F25E]HBP, A MUTANT OF HUMAN HEPARIN BINDING PROTEIN (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHANOL, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
1FY3
 
 | | [G175Q]HBP, A mutant of human heparin binding protein (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
1TN3
 
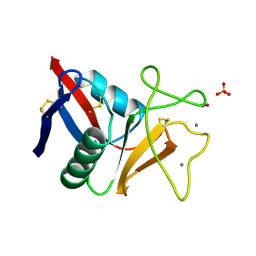 | | THE C-TYPE LECTIN CARBOHYDRATE RECOGNITION DOMAIN OF HUMAN TETRANECTIN | | Descriptor: | CALCIUM ION, ETHANOL, SULFATE ION, ... | | Authors: | Kastrup, J.S, Nielsen, B.B, Rasmussen, H, Holtet, T.L, Graversen, J.H, Etzerodt, M, Thoegersen, H.C, Larsen, I.K. | | Deposit date: | 1997-11-06 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-type lectin carbohydrate recognition domain of human tetranectin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1N0T
 
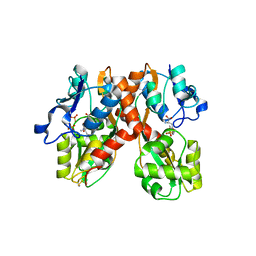 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
1EPF
 
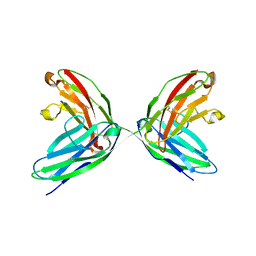 | | CRYSTAL STRUCTURE OF THE TWO N-TERMINAL IMMUNOGLOBULIN DOMAINS OF THE NEURAL CELL ADHESION MOLECULE (NCAM) | | Descriptor: | CALCIUM ION, PROTEIN (NEURAL CELL ADHESION MOLECULE) | | Authors: | Kasper, C, Rasmussen, H, Kastrup, J.S, Ikemizu, S, Jones, E.Y, Berezin, V, Bock, E, Larsen, I.K. | | Deposit date: | 2000-03-29 | | Release date: | 2000-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of cell-cell adhesion by NCAM.
Nat.Struct.Biol., 7, 2000
|
|
1XJ9
 
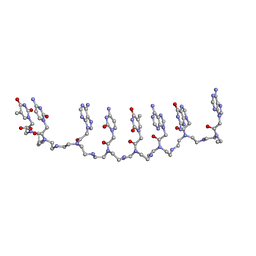 | | Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network | | Descriptor: | peptide nucleic acid, (H-P(*GPN*TPN*APN*GPN*APN*TPN*CPN*APN*CPN*TPN)-LYS-NH2) | | Authors: | Petersson, B, Nielsen, B.B, Rasmussen, H, Larsen, I.K, Gajhede, M, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2004-09-23 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Partly Self-Complementary Peptide Nucleic Acid (PNA) Oligomer Showing a Duplex-Triplex Network
J.Am.Chem.Soc., 127, 2005
|
|
1HTN
 
 | | HUMAN TETRANECTIN, A TRIMERIC PLASMINOGEN BINDING PROTEIN WITH AN ALPHA-HELICAL COILED COIL | | Descriptor: | CALCIUM ION, TETRANECTIN | | Authors: | Nielsen, B.B, Kastrup, J.S, Rasmussen, H, Holtet, T.L, Graversen, J.H, Etzerodt, M, Thogersen, H.C, Larsen, I.K. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of tetranectin, a trimeric plasminogen-binding protein with an alpha-helical coiled coil.
FEBS Lett., 412, 1997
|
|
1M5C
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5D
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
3PD8
 
 | | X-ray structure of the ligand-binding core of GluA2 in complex with (S)-7-HPCA at 2.5 A resolution | | Descriptor: | (7S)-3-hydroxy-4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridine-7-carboxylic acid, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Biostructural and pharmacological studies of bicyclic analogues of the 3-isoxazolol glutamate receptor agonist ibotenic acid.
J. Med. Chem., 53, 2010
|
|
1XSM
 
 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
2P2A
 
 | | X-ray structure of the GluR2 ligand binding core (S1S2J) in complex with 2-Bn-tet-AMPA at 2.26A resolution | | Descriptor: | 2-AMINO-3-[3-HYDROXY-5-(2-BENZYL-2H-5-TETRAZOLYL)-4-ISOXAZOLYL]-PROPIONIC ACID, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Frydenvang, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A tetrazolyl-substituted subtype-selective AMPA receptor agonist.
J.Med.Chem., 50, 2007
|
|
1VSO
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
1WVJ
 
 | | Exploring the GluR2 ligand-binding core in complex with the bicyclic AMPA analogue (S)-4-AHCP | | Descriptor: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, GLYCEROL, SULFATE ION, ... | | Authors: | Nielsen, B.B, Pickering, D.S, Greenwood, J.R, Brehm, L, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the GluR2 ligand-binding core in complex with the bicyclical AMPA analogue (S)-4-AHCP
FEBS J., 272, 2005
|
|
