4CVY
 
 | | Crystal structure of the M. tuberculosis sulfate ester dioxygenase Rv3406 in complex with iron. | | Descriptor: | DIOXYGENASE RV3406/MT3514, FE (III) ION, NITRATE ION | | Authors: | Neres, J, Hartkoorn, R.C, Chiarelli, L.R, Gadupudi, R, Pasca, M, Mori, G, Farina, D, Salina, S, Makarov, V, Kolly, G.S, Molteni, E, Binda, C, Dhar, N, Ferrari, S, Brodin, P, Delorme, V, Landry, V, de Jesus Lopes Ribeiro, A.L, Saxena, P, Pojer, F, Venturelli, A, Carta, A, Luciani, R, Porta, A, Zanoni, G, De Rossi, E, Costi, M.P, Riccardi, G, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium Tuberculosis Through Noncovalent Inhibition of Dpre1.
Acs Chem.Biol., 10, 2015
|
|
4QIA
 
 | | Crystal structure of human insulin degrading enzyme (ide) in complex with inhibitor N-benzyl-N-(carboxymethyl)glycyl-L-histidine | | Descriptor: | Insulin-degrading enzyme, N-benzyl-N-(carboxymethyl)glycyl-L-histidine, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
2YPU
 
 | | human insulin degrading enzyme E111Q in complex with inhibitor compound 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, INSULIN-DEGRADING ENZYME, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.-J. | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Imidazole-Derived 2-[N-Carbamoylmethyl-Alkylamino]Acetic Acids,Substrate-Dependent Modulators of Insulin-Degrading Enzyme in Amyloid-Beta Hydrolysis
Eur J Med Chem, 79C, 2014
|
|
8AEB
 
 | | SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(pyridin-3-ylmethyl)thioformamide, ... | | Authors: | Hanoulle, X, Charton, J, Deprez, B. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
4RE9
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4P8T
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor QN129 | | Descriptor: | (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 3-[(4-cyanobenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
4P8N
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor QN118 | | Descriptor: | 3-[(3-fluoro-4-methoxybenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
4P8K
 
 | |
4P8L
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor Ty36c | | Descriptor: | 3-[(4-fluorobenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
4P8C
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor QN127 | | Descriptor: | (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 6-(trifluoromethyl)-3-{[4-(trifluoromethyl)benzyl]amino}quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
4P8M
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor QN114 | | Descriptor: | (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 3-[(4-ethoxybenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
4P8P
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor QN127 | | Descriptor: | (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 3-[(4-chlorobenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
4P8Y
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor Ty21c | | Descriptor: | 3-[(4-methoxybenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
3QZ2
 
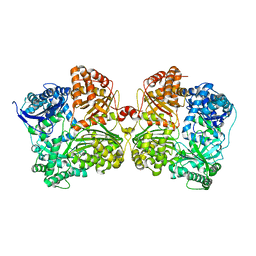 | | The structure of cysteine-free human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme, ZINC ION | | Authors: | Guo, Q, Tang, W.J. | | Deposit date: | 2011-03-04 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
7NTS
 
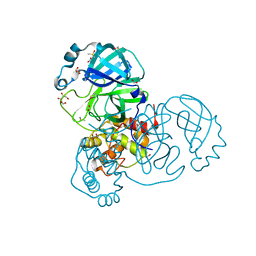 | | Crystal structure of the SARS-CoV-2 Main Protease with oxidized C145 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NTQ
 
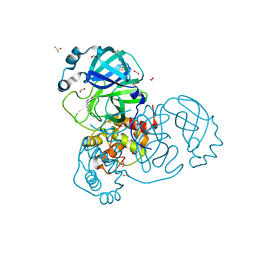 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
4DWK
 
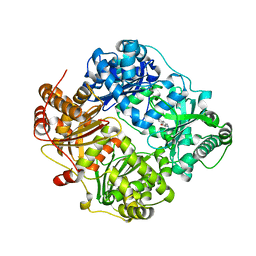 | | Structure of cystein free insulin degrading enzyme with compound bdm41671 ((s)-2-{2-[carboxymethyl-(3-phenyl-propyl)-amino]-acetylamino}-3-(1h-imidazol-4-yl)-propionic acid methyl ester) | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-L-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-02-24 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4DTT
 
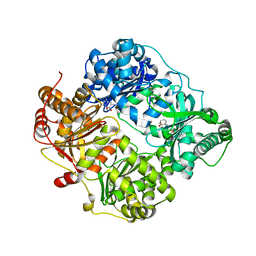 | | Crystal structure of human insulin degrading enzyme (ide) in complex with compund 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, Insulin-degrading enzyme, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-02-21 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4GS8
 
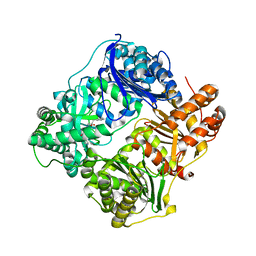 | | Structure analysis of cysteine free insulin degrading enzyme (ide) with compound bdm43079 [{[(s)-2-(1h-imidazol-4-yl)-1-methylcarbamoyl-ethylcarbamoyl]-methyl}-(3-phenyl-propyl)-amino]-acetic acid | | Descriptor: | Insulin-degrading enzyme, N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-N-methyl-L-histidinamide, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4GSC
 
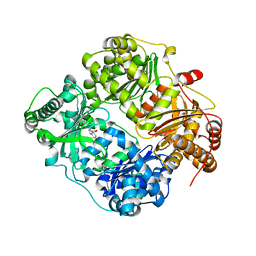 | | Structure analysis of insulin degrading enzyme with compound bdm41559 ((s)-2-[2-(carboxymethyl-phenethyl-amino)-acetylamino]-3-(1h-imidazol-4-yl)-propionic acid methyl ester) | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(2-phenylethyl)glycyl-L-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4GSF
 
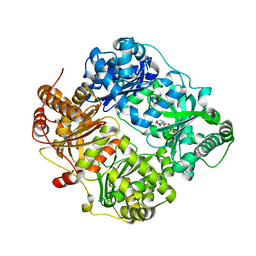 | | The structure analysis of cysteine free insulin degrading enzyme (ide) with (s)-2-{2-[carboxymethyl-(3-phenyl-propionyl)-amino]-acetylamino}-3-(3h-imidazol-4-yl)-propionic acid methyl ester | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropanoyl)glycyl-D-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
7P51
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 MAIN PROTEASE COMPLEXED WITH FRAGMENT F01 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide, ... | | Authors: | Hanoulle, X, Moschidi, D. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4NXO
 
 | | Crystal Structure of Insulin Degrading Enzyme in complex with BDM44768 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2013-12-09 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4IFH
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 | | Descriptor: | Insulin-degrading enzyme, N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)-4-methylbenzamide, ZINC ION | | Authors: | Liang, W.G, Guo, Q, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
