6O6C
 
 | | RNA polymerase II elongation complex arrested at a CPD lesion | | Descriptor: | DNA (27-MER), DNA (5'-D(P*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*CP*AP*GP*AP*GP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leshziner, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | 3.1 angstrom structure of yeast RNA polymerase II elongation complex stalled at a cyclobutane pyrimidine dimer lesion solved using streptavidin affinity grids.
J.Struct.Biol., 207, 2019
|
|
7MGM
 
 | | Structure of yeast cytoplasmic dynein with AAA3 Walker B mutation bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lahiri, I, Reimer, J.M, Leschziner, A.E. | | Deposit date: | 2021-04-12 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for cytoplasmic dynein-1 regulation by Lis1.
Elife, 11, 2022
|
|
5VVR
 
 | |
5VVS
 
 | | RNA pol II elongation complex | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leschziner, A.E. | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structural basis for the initiation of eukaryotic transcription-coupled DNA repair.
Nature, 551, 2017
|
|
8DZZ
 
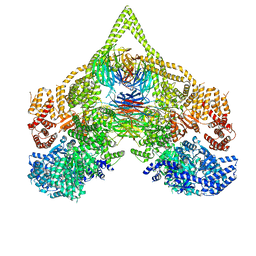 | | Cryo-EM structure of chi dynein bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E00
 
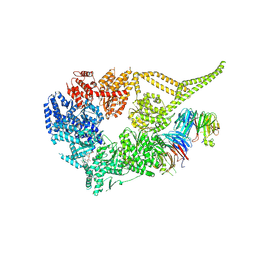 | | Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6DRV
 
 | |
7YL9
 
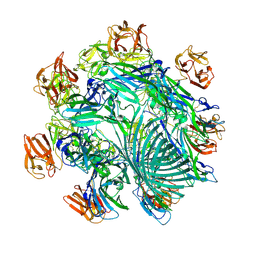 | | Cryo-EM structure of complete transmembrane channel E289A mutant Vibrio cholerae Cytolysin | | Descriptor: | Hemolysin | | Authors: | Mondal, A.K, Sengupta, N, Singh, M, Biswas, R, Lata, K, Lahiri, I, Dutta, S, Chattopadhyay, K. | | Deposit date: | 2022-07-25 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of complete transmembrane channel E289A mutant Vibrio cholerae Cytolysin
J.Biol.Chem.
|
|
6VNO
 
 | |
6VP8
 
 | |
6VP6
 
 | |
6VP7
 
 | |
7LGM
 
 | | Cyanophycin synthetase from A. baylyi DSM587 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase | | Authors: | Sharon, I, Haque, A.S, Lahiri, I, Leschziner, A, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
8TVS
 
 | | Cryo-EM structure of backtracked Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVQ
 
 | | Cryo-EM structure of CPD stalled 10-subunit Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVP
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TUG
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (engaged state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVV
 
 | | Cryo-EM structure of backtracked Pol II | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVX
 
 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVW
 
 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7LGQ
 
 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 8x(Asp-Arg)-Asn | | Descriptor: | 8x(Asp-Arg)-Asn, ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
7LGN
 
 | |
7LG5
 
 | |
7LGJ
 
 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 8x(Asp-Arg)-NH2 | | Descriptor: | 8x(Asp-Arg)-NH2, Cyanophycin synthase, MAGNESIUM ION, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and function of the amino acid polymerase cyanophycin synthetase.
Nat.Chem.Biol., 17, 2021
|
|
