148L
 
 | | A COVALENT ENZYME-SUBSTRATE INTERMEDIATE WITH SACCHARIDE DISTORTION IN A MUTANT T4 LYSOZYME | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, BETA-MERCAPTOETHANOL, SUBSTRATE CLEAVED FROM CELL WALL OF ESCHERICHIA COLI, ... | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1993-10-27 | | Release date: | 1994-04-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A covalent enzyme-substrate intermediate with saccharide distortion in a mutant T4 lysozyme.
Science, 262, 1993
|
|
1I20
 
 | | MUTANT HUMAN LYSOZYME (A92D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I1Z
 
 | | MUTANT HUMAN LYSOZYME (Q86D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I22
 
 | | MUTANT HUMAN LYSOZYME (A83K/Q86D/A92D) | | Descriptor: | CALCIUM ION, LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1QTZ
 
 | | D20C MUTANT OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT3
 
 | | T26D MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (T4 Lysozyme) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QTV
 
 | | T26E APO STRUCTURE OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT8
 
 | | T26H Mutant of T4 Lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT7
 
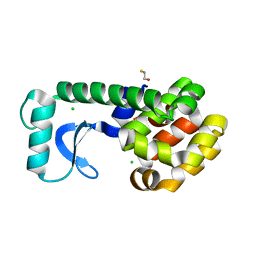 | | E11N Mutant of T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT4
 
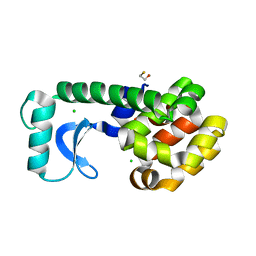 | | T26Q MUTANT OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT6
 
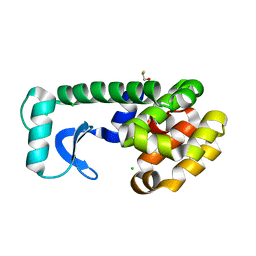 | | E11H Mutant of T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QT5
 
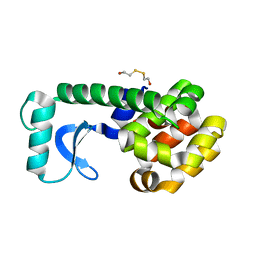 | | D20E MUTANT STRUCTURE OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, PROTEIN (T4 LYSOZYME) | | Authors: | Kuroki, R, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1999-06-30 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1LZ4
 
 | | ENTHALPIC DESTABILIZATION OF A MUTANT HUMAN LYSOZYME LACKING A DISULFIDE BRIDGE BETWEEN CYSTEINE-77 AND CYSTEINE-95 | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Kuroki, R, Taniyama, Y, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enthalpic destabilization of a mutant human lysozyme lacking a disulfide bridge between cysteine-77 and cysteine-95.
Biochemistry, 31, 1992
|
|
254L
 
 | | LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1997-11-10 | | Release date: | 1998-01-28 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
253L
 
 | | LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1997-11-10 | | Release date: | 1998-01-28 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
255L
 
 | | HYDROLASE | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1997-11-10 | | Release date: | 1998-01-28 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1CD9
 
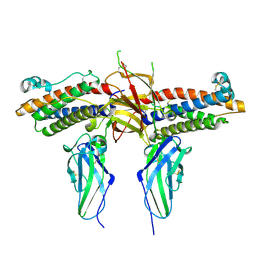 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
4WHM
 
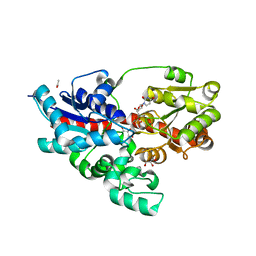 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
3OTJ
 
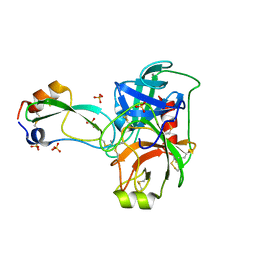 | | A Crystal Structure of Trypsin Complexed with BPTI (Bovine Pancreatic Trypsin Inhibitor) by X-ray/Neutron Joint Refinement | | Descriptor: | CALCIUM ION, Cationic trypsin, Pancreatic trypsin inhibitor, ... | | Authors: | Kawamura, K, Yamada, T, Kurihara, K, Tamada, T, Kuroki, R, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2010-09-12 | | Release date: | 2011-01-26 | | Last modified: | 2017-11-08 | | Method: | NEUTRON DIFFRACTION (2.15 Å), X-RAY DIFFRACTION | | Cite: | X-ray and neutron protein crystallographic analysis of the trypsin-BPTI complex.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4REN
 
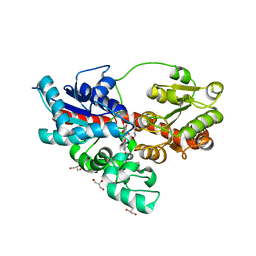 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | Descriptor: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REL
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
3HGN
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by neutron crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.65 Å), X-RAY DIFFRACTION | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
1EHA
 
 | | CRYSTAL STRUCTURE OF GLYCOSYLTREHALOSE TREHALOHYDROLASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
1EH9
 
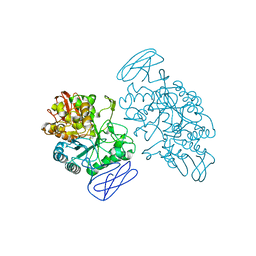 | | CRYSTAL STRUCTURE OF SULFOLOBUS SOLFATARICUS GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
