3WZ4
 
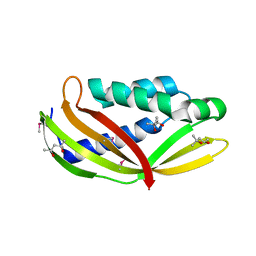 | | Structure of the periplasmic domain of DotI (crystal form I) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DotI | | Authors: | Kuroda, T, Kubori, T, Uchida, Y, Nagai, H, Imada, K. | | Deposit date: | 2014-09-18 | | Release date: | 2015-06-10 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion
Sci Rep, 5, 2015
|
|
3WZ3
 
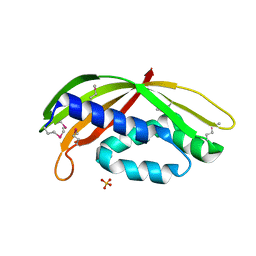 | | Structure of a periplasmic fragment of TraM | | Descriptor: | SULFATE ION, TraM protein | | Authors: | Kuroda, T, Kubori, T, Uchida, Y, Nagai, H, Imada, K. | | Deposit date: | 2014-09-18 | | Release date: | 2015-06-10 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion
Sci Rep, 5, 2015
|
|
3WZ5
 
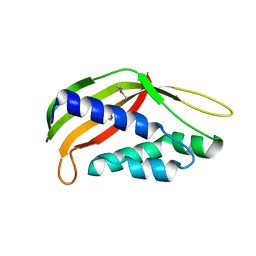 | | Structure of the periplasmic domain of DotI (crystal form II) | | Descriptor: | DotI | | Authors: | Kuroda, T, Kubori, T, Uchida, Y, Nagai, H, Imada, K. | | Deposit date: | 2014-09-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion
Sci Rep, 5, 2015
|
|
6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IDP
 
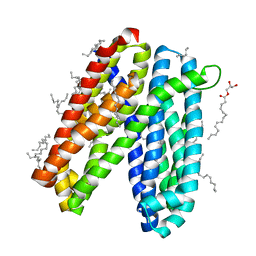 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the straight form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IDR
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
1WUA
 
 | | The structure of Aplyronine A-actin complex | | Descriptor: | (8R,9R,10R,11R,14S,18S,20S,24S)-24-{(1R,2S,3R,6R,7R,8R,9S,10E)-8-(ACETYLOXY)-6-[(N,N-DIMETHYLALANYL)OXY]-11-[FORMYL(MET HYL)AMINO]-2-HYDROXY-1,3,7,9-TETRAMETHYLUNDEC-10-ENYL}-10-HYDROXY-14,20-DIMETHOXY-9,11,15,18-TETRAMETHYL-2-OXOOXACYCLOTE TRACOSA-3,5,15,21-TETRAEN-8-YL N,N,O-TRIMETHYLSERINATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Hirata, K, Muraoka, S, Suenaga, K, Kuroda, T, Kato, K, Tanaka, H, Yamamoto, M, Takata, M, Yamada, K, Kigoshi, H. | | Deposit date: | 2004-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure basis for antitumor effect of aplyronine a
J.Mol.Biol., 356, 2006
|
|
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
3VVP
 
 | | Crystal structure of MATE in complex with Br-NRF | | Descriptor: | 6-bromo-1-ethyl-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3VVR
 
 | | Crystal structure of MATE in complex with MaD5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3VVS
 
 | | Crystal structure of MATE in complex with MaD3S | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3VVN
 
 | |
3VVO
 
 | |
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3W4T
 
 | | Crystal structure of MATE P26A mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-01-16 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
6LUH
 
 | | High resolution structure of N(omega)-hydroxy-L-arginine hydrolase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, N(omega)-hydroxy-L-arginine amidinohydrolase | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an Nomega-hydroxy-L-arginine hydrolase found in the D-cycloserine biosynthetic pathway.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6LUG
 
 | | Crystal structure of N(omega)-hydroxy-L-arginine hydrolase | | Descriptor: | MANGANESE (II) ION, N(omega)-hydroxy-L-arginine amidinohydrolase | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an Nomega-hydroxy-L-arginine hydrolase found in the D-cycloserine biosynthetic pathway.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
