8H7C
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) - Pt derivative | | Descriptor: | ACETATE ION, CHLORIDE ION, De novo ferric enterobactin esterase Syn-F4, ... | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H7E
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) at 2.0 angstrom resolution | | Descriptor: | ACETATE ION, De novo ferric enterobactin esterase Syn-F4 | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H7D
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) | | Descriptor: | ACETATE ION, De novo ferric enterobactin esterase Syn-F4 | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1VCX
 
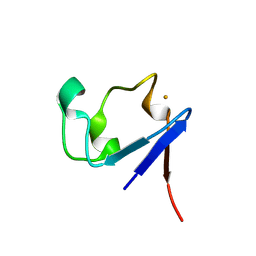 | | Neutron Crystal Structure of the Wild Type Rubredoxin from Pyrococcus Furiosus at 1.5A Resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Kurihara, K, Tanaka, I, Chatake, T, Adams, M.W.W, Jenney Jr, F.E, Moiseeva, N, Bau, R, Niimura, N. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Neutron crystallographic study on rubredoxin from Pyrococcus furiosus by BIX-3, a single-crystal diffractometer for biomacromolecules
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3OTJ
 
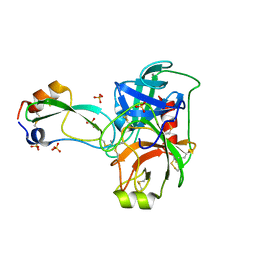 | | A Crystal Structure of Trypsin Complexed with BPTI (Bovine Pancreatic Trypsin Inhibitor) by X-ray/Neutron Joint Refinement | | Descriptor: | CALCIUM ION, Cationic trypsin, Pancreatic trypsin inhibitor, ... | | Authors: | Kawamura, K, Yamada, T, Kurihara, K, Tamada, T, Kuroki, R, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2010-09-12 | | Release date: | 2011-01-26 | | Last modified: | 2017-11-08 | | Method: | NEUTRON DIFFRACTION (2.15 Å), X-RAY DIFFRACTION | | Cite: | X-ray and neutron protein crystallographic analysis of the trypsin-BPTI complex.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4GPG
 
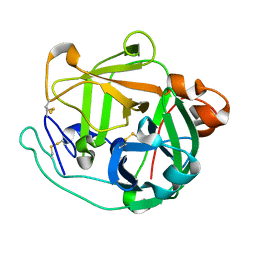 | | X/N joint refinement of Achromobacter Lyticus Protease I free form at pD8.0 | | Descriptor: | Protease 1 | | Authors: | Ohnishi, Y, Yamada, T, Kurihara, K, Tanaka, I, Sakiyama, F, Masaki, T, Niimura, N. | | Deposit date: | 2012-08-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | NEUTRON DIFFRACTION (1.895 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of Achromobacter protease I at pD 8.0: protonation states and hydration structure in the free-form.
Biochim.Biophys.Acta, 1834, 2013
|
|
8K9N
 
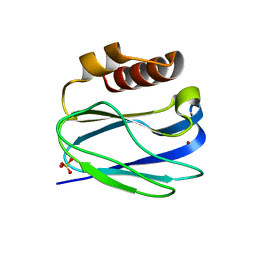 | | Subatomic resolution structure of Pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Lintuluoto, M, Kurihara, K, Hasegawa, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
8K9P
 
 | | Neutron X-ray joint structure of pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Kurihara, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3U2J
 
 | | Neutron crystal structure of human Transthyretin | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3U2I
 
 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3VXF
 
 | | X/N Joint refinement of Human alpha-thrombin-Bivalirudin complex PD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.602 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
3VXE
 
 | | Human alpha-thrombin-Bivalirudin complex at PD5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
3WA3
 
 | | Crystal structure of copper amine oxidase from arthrobacter globiformis in N2 condition | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WA2
 
 | | High resolution crystal structure of copper amine oxidase from arthrobacter globiformis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FHP
 
 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | Descriptor: | Insulin, ZINC ION | | Authors: | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1IU5
 
 | | X-ray Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IU6
 
 | | Neutron Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3VTE
 
 | | Crystal structure of tetrahydrocannabinolic acid synthase from Cannabis sativa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Tetrahydrocannabinolic acid synthase | | Authors: | Shoyama, Y, Tamada, T, Kurihara, K, Takeuchi, A, Taura, F, Arai, S, Blaber, M, Shoyama, Y, Morimoto, S, Kuroki, R. | | Deposit date: | 2012-05-28 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of 1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa
J.Mol.Biol., 423, 2012
|
|
1WQZ
 
 | | Complicated water orientations in the minor groove of B-DNA decamer D(CCATTAATGG)2 observed by neutron diffraction measurements | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (3 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WQY
 
 | | X-RAY structural analysis of B-DNA decamer D(CCATTAATGG)2 crystal grown in D2O solution | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
8W6X
 
 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
3HGN
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by neutron crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.65 Å), X-RAY DIFFRACTION | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
3FX5
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
8J6G
 
 | |
