5WXU
 
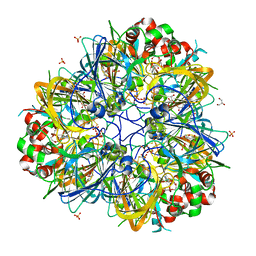 | | 11S globulin from Wrightia tinctoria reveals auxin binding site | | Descriptor: | 11S globulin, 1H-INDOL-3-YLACETIC ACID, CITRATE ANION, ... | | Authors: | Kumar, P, Kesari, P, Dhindwal, S, Kumar, P. | | Deposit date: | 2017-01-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel function for globulin in sequestering plant hormone: Crystal structure of Wrightia tinctoria 11S globulin in complex with auxin.
Sci Rep, 7, 2017
|
|
6A80
 
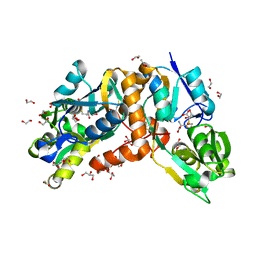 | | Crystal Structure of putative amino acid binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus in complex with cystine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-05 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
6A8S
 
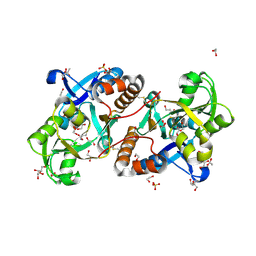 | | Crystal Structure of the putative amino acid-binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus in complex with Cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-10 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
6AA1
 
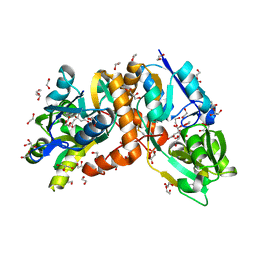 | | Crystal structure of putative amino acid binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus bound with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-16 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
6AAL
 
 | | Crystal Structure of putative amino acid binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus in complex with Arginine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-18 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
4R62
 
 | | Structure of Rad6~Ub | | Descriptor: | ACETATE ION, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 2 | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Role of a non-canonical surface of Rad6 in ubiquitin conjugating activity.
Nucleic Acids Res., 43, 2015
|
|
6IXI
 
 | | structure of Cd-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | Authors: | Kumar, P, Sharma, N, Dalal, V, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-12-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of the heavy metal binding properties of periplasmic metal uptake protein CLas-ZnuA2.
Metallomics, 12, 2020
|
|
6NP5
 
 | | AAC-VIa bound to Kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.353 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NP4
 
 | | AAC-VIa bound to Tobramycin | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION, TOBRAMYCIN | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NP1
 
 | |
6NP3
 
 | | AAC-VIa bound to Gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6NP2
 
 | | AAC-VIa bound to Sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6O5U
 
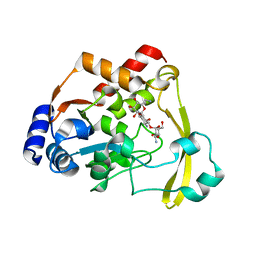 | | AAC-VIa bound to Kanamycin A | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, KANAMYCIN A, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-03-04 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7QDJ
 
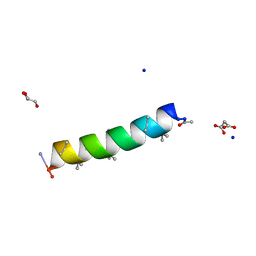 | | Racemic structure of PK-10 and PK-11 | | Descriptor: | GLYCEROL, MALONATE ION, PK-10+PK-11, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7QDK
 
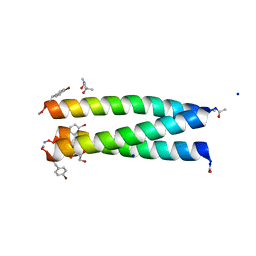 | | A trimeric de novo coiled-coil assembly: CC-TypeN-LaLd | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CC-TypeN-LaLd, GLYCEROL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
7QDI
 
 | | Structure of octameric left-handed 310-helix bundle: D-310HD | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2021-11-27 | | Release date: | 2022-04-27 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
3BVN
 
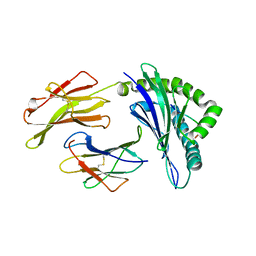 | | High resolution crystal structure of HLA-B*1402 in complex with the latent membrane protein 2 peptide (LMP2) of Epstein-Barr virus | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B*1402 alpha chain, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2008-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
4R7E
 
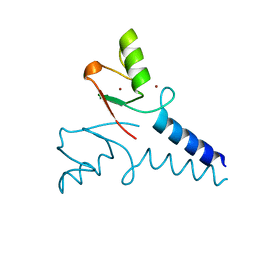 | | Structure of Bre1 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1, ZINC ION | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of the yeast Bre1 RING domain.
Proteins, 83, 2015
|
|
8GYR
 
 | | Crystal structure of a variable region segment of Leptospira host-interacting outer surface protein, LigA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kumar, P, Akif, M. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of a variable region segment of Leptospira host-interacting outer surface protein, LigA, reveals the orientation of Ig-like domains.
Int.J.Biol.Macromol., 244, 2023
|
|
3BO8
 
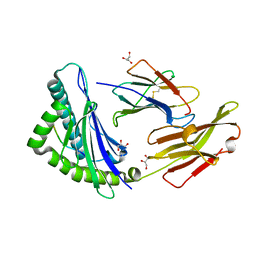 | | The High Resolution Crystal Structure of HLA-A1 Complexed with the MAGE-A1 Peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational changes within the HLA-A1:MAGE-A1 complex induced by binding of a recombinant antibody fragment with TCR-like specificity
Protein Sci., 18, 2009
|
|
5E33
 
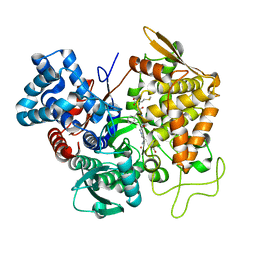 | | Structure of human DPP3 in complex with met-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, Met-enkephalin, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5E3A
 
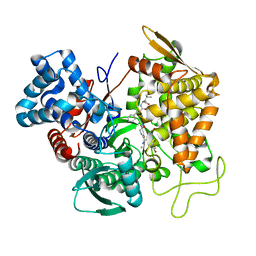 | | Structure of human DPP3 in complex with opioid peptide leu-enkephalin | | Descriptor: | Dipeptidyl peptidase 3, Leu-enkephalin, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5E3C
 
 | | Structure of human DPP3 in complex with hemorphin like opioid peptide IVYPW | | Descriptor: | Dipeptidyl peptidase 3, IVYPW, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5EGY
 
 | | Structure of ligand free human DPP3 in closed form. | | Descriptor: | Dipeptidyl peptidase 3, MAGNESIUM ION, ZINC ION | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.741 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
5EHH
 
 | | Structure of human DPP3 in complex with endomorphin-2. | | Descriptor: | Dipeptidyl peptidase 3, Endomorphin-2, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Reisinger, M, Pavkov-Keller, T, Wallner, S, Macheroux, P, Gruber, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition.
Sci Rep, 6, 2016
|
|
