1TV3
 
 | | Crystal structure of the N-methyl-hydroxylamine MtmB complex | | Descriptor: | 5-(HYDROXY-METHYL-AMINO)-3-METHYL-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
1TV2
 
 | | Crystal structure of the hydroxylamine MtmB complex | | Descriptor: | 5-HYDROXYAMINO-3-METHYL-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
1TV4
 
 | | Crystal structure of the sulfite MtmB complex | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1, SULFATE ION | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
1L2Q
 
 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
1NTH
 
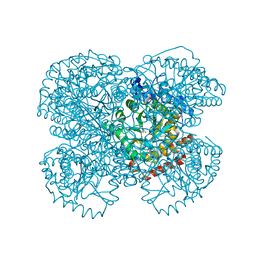 | | Crystal structure of the methanosarcina barkeri monomethylamine methyltransferase (MTMB) | | Descriptor: | Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase
Science, 296, 2002
|
|
3CF4
 
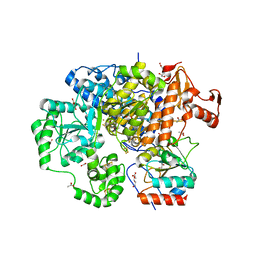 | | Structure of the CODH component of the M. barkeri ACDS complex | | Descriptor: | ACETIC ACID, Acetyl-CoA decarboxylase/synthase alpha subunit, Acetyl-CoA decarboxylase/synthase epsilon subunit, ... | | Authors: | Gong, W, Hao, B, Wei, Z, Ferguson Jr, D.J, Tallant, T, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2008-03-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the alpha2 epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarboxylase/synthase complex
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7XCM
 
 | | Crystal structure of sulfite MttB structure at 3.2 A resolution | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7XCL
 
 | |
3EZX
 
 | |
