4WWL
 
 | | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form) | | Descriptor: | CARBONATE ION, GLYCEROL, Protein UshA, ... | | Authors: | Krug, U, Paithankar, K.S, Schultz-Heienbrok, R, Strater, N. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form)
To Be Published
|
|
4A5B
 
 | |
4A57
 
 | |
4A59
 
 | |
4A5A
 
 | |
4JEP
 
 | |
4KH5
 
 | | Toxoplasma gondii NTPDase1 C258S/C268S in complex with Mg and AMPNP | | Descriptor: | 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, MAGNESIUM ION, Nucleoside-triphosphatase 2 | | Authors: | Krug, U, Totzauer, R, Strater, N. | | Deposit date: | 2013-04-30 | | Release date: | 2013-11-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The ATP/ADP substrate specificity switch between Toxoplasma gondii NTPDase1 and NTPDase3 is caused by an altered mode of binding of the substrate base.
Chembiochem, 14, 2013
|
|
4KH4
 
 | | Toxoplasma gondii NTPDase1 C258S/C268S in complex with Mg and AMPPNP | | Descriptor: | MAGNESIUM ION, Nucleoside-triphosphatase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Krug, U, Totzauer, R, Strater, N. | | Deposit date: | 2013-04-30 | | Release date: | 2013-11-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The ATP/ADP substrate specificity switch between Toxoplasma gondii NTPDase1 and NTPDase3 is caused by an altered mode of binding of the substrate base.
Chembiochem, 14, 2013
|
|
4KH6
 
 | |
6HDX
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with R3-HIB-AMP | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] (2~{R})-2-methyl-3-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HE2
 
 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HE0
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA in the thioesterfication state | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HDW
 
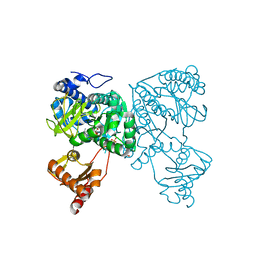 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with 2-HIB-AMP | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] 2-methyl-2-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HDY
 
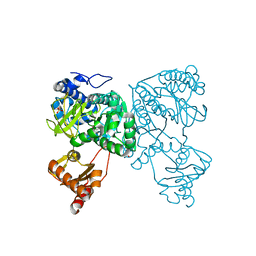 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with S3-HB-AMP | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
