3M3G
 
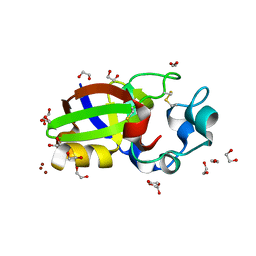 | | Crystal structure of Sm1, an elicitor of plant defence responses from Trichoderma virens. | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Epl1 protein, ... | | Authors: | Krieger, I.V, Vargas, W.A, Kenerley, C.M, Sacchettini, J.C. | | Deposit date: | 2010-03-09 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of Sm1, an elicitor of plant defence responses from Trichoderma virens.
To be Published
|
|
6PXE
 
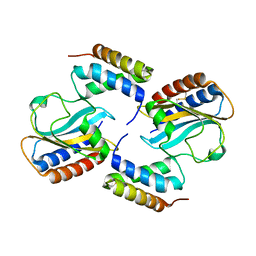 | |
6PX4
 
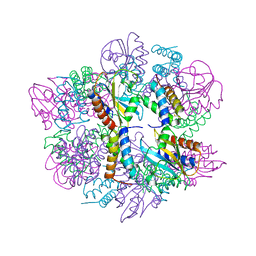 | |
3S9I
 
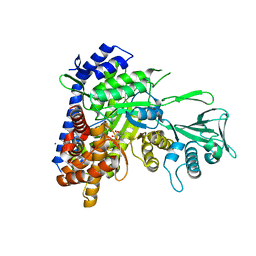 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 2-4-dioxo-4-phenylbutanoic acid inhibitor | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sun, Q, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2011-06-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase.
Chem.Biol., 19, 2012
|
|
3S9Z
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 4-(2-bromophenyl)-2,4-dioxobutanoic acid inhibitor | | Descriptor: | 4-(2-bromophenyl)-2,4-dioxobutanoic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sun, Q, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2011-06-02 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase.
Chem.Biol., 19, 2012
|
|
3SAD
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 4-(2-mehtylphenyl)-2,4-dioxobutanoic acid inhibitor | | Descriptor: | 4-(2-methylphenyl)-2,4-dioxobutanoic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sun, Q, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2011-06-02 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase.
Chem.Biol., 19, 2012
|
|
3SAZ
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 4-(3-bromophenyl)-2,4-dioxobutanoic acid inhibitor | | Descriptor: | 4-(3-bromophenyl)-2,4-dioxobutanoic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sun, Q, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2011-06-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase.
Chem.Biol., 19, 2012
|
|
3SB0
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 4-(2-chloro-6-fluoro-3-methylphenyl)-2,4-dioxobutanoic acid inhibitor | | Descriptor: | 4-(2-chloro-6-fluoro-3-methylphenyl)-2,4-dioxobutanoic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sun, Q, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2011-06-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase.
Chem.Biol., 19, 2012
|
|
6MJ1
 
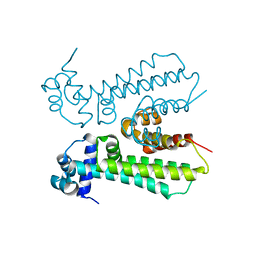 | |
8SUT
 
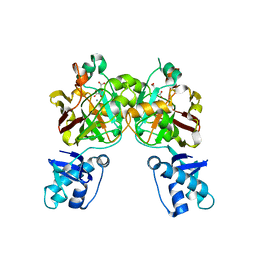 | | Crystal structure of YisK from Bacillus subtilis in complex with reaction product 4-Hydroxy-2-oxoglutaric acid | | Descriptor: | (2~{R})-2-oxidanyl-4-oxidanylidene-pentanedioic acid, Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION | | Authors: | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
8SUU
 
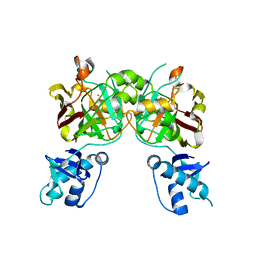 | | Crystal structure of YisK from Bacillus subtilis in apo form | | Descriptor: | Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION | | Authors: | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
8SKY
 
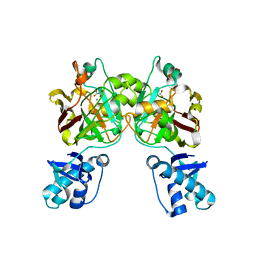 | | Crystal structure of YisK from Bacillus subtilis in complex with oxalate | | Descriptor: | Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION, OXALATE ION | | Authors: | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | Deposit date: | 2023-04-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
7RB1
 
 | | Isocitrate Lyase-1 from Mycobacterium tuberculosis covalently modified by 5-descarboxy-5-nitro-D-isocitric acid | | Descriptor: | (3E)-3-(hydroxyimino)propanoic acid, GLYCEROL, GLYOXYLIC ACID, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T, Sacchettini, J.C. | | Deposit date: | 2021-07-05 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Based Inactivation of Mycobacterium tuberculosis Isocitrate Lyase 1 by (2 R ,3 S )-2-Hydroxy-3-(nitromethyl)succinic acid.
J.Am.Chem.Soc., 143, 2021
|
|
7SVT
 
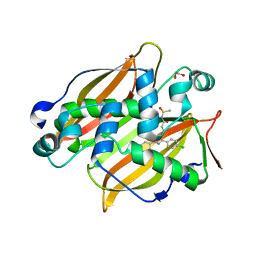 | | Mycobacterium tuberculosis 3-hydroxyl-ACP dehydratase HadAB in complex with 1,3-diarylpyrazolyl-acylsulfonamide inhibitor | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 1,2-ETHANEDIOL, 3-[1-(4-bromophenyl)-3-(4-chlorophenyl)-1H-pyrazol-4-yl]-N-(methanesulfonyl)propanamide, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1,3-Diarylpyrazolyl-acylsulfonamides Target HadAB/BC Complex in Mycobacterium tuberculosis .
Acs Infect Dis., 8, 2022
|
|
5H8U
 
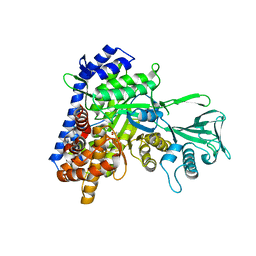 | | Crystal structure of mycobacterium tuberculosis wild-type malate synthase in complex with product malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Krieger, I.V, Huang, H.-L, Sacchettini, J.C. | | Deposit date: | 2015-12-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mycobacterium tuberculosis Malate Synthase Structures with Fragments Reveal a Portal for Substrate/Product Exchange.
J. Biol. Chem., 291, 2016
|
|
6PSH
 
 | |
6PSK
 
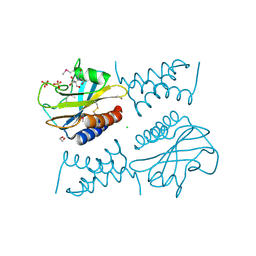 | | Crystal structure of the complex between periplasmic domains of antiholin RI and holin T from T4 phage, in P6522 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Antiholin, ... | | Authors: | Kuznetsov, V.B, Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition.
J.Mol.Biol., 432, 2020
|
|
6DKO
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 2,6-F-phenyldiketoacid | | Descriptor: | 4-(2,6-difluorophenyl)-2,4-dioxobutanoic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Anion-pi Interactions in Computer-Aided Drug Design: Modeling the Inhibition of Malate Synthase by Phenyl-Diketo Acids.
J Chem Inf Model, 58, 2018
|
|
6DNP
 
 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 2-F-3-Methyl-6-F-phenyldiketoacid | | Descriptor: | (2Z)-4-(2,6-difluoro-3-methylphenyl)-2-hydroxy-4-oxobut-2-enoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC), Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Anion-pi Interactions in Computer-Aided Drug Design: Modeling the Inhibition of Malate Synthase by Phenyl-Diketo Acids.
J Chem Inf Model, 58, 2018
|
|
5DRI
 
 | |
5ECV
 
 | |
5H8P
 
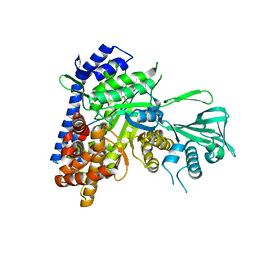 | |
5H8M
 
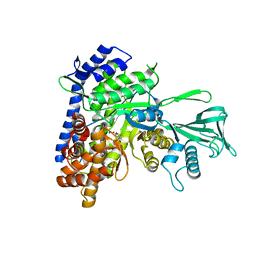 | | Crystal structure of Mycobacterium tuberculosis malate synthase C619A, G459A mutant in complex with product malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MAGNESIUM ION, Malate synthase G | | Authors: | Krieger, I.V, Huang, H.-L, Sacchettini, J.C. | | Deposit date: | 2015-12-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis Malate Synthase Structures with Fragments Reveal a Portal for Substrate/Product Exchange.
J. Biol. Chem., 291, 2016
|
|
6AU9
 
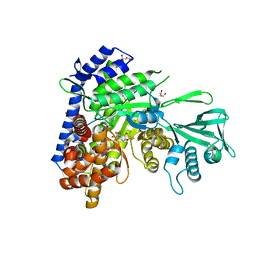 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with dioxine-phenyldiketoacid | | Descriptor: | (2Z)-4-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-hydroxy-4-oxobut-2-enoic acid, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2017-08-31 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anion-pi Interactions in Computer-Aided Drug Design: Modeling the Inhibition of Malate Synthase by Phenyl-Diketo Acids.
J Chem Inf Model, 58, 2018
|
|
6APZ
 
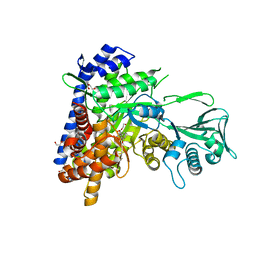 | |
