1AGT
 
 | | SOLUTION STRUCTURE OF THE POTASSIUM CHANNEL INHIBITOR AGITOXIN 2: CALIPER FOR PROBING CHANNEL GEOMETRY | | Descriptor: | AGITOXIN 2 | | Authors: | Krezel, A.M, Kasibhatla, C, Hidalgo, P, Mackinnon, R, Wagner, G. | | Deposit date: | 1995-04-14 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the potassium channel inhibitor agitoxin 2: caliper for probing channel geometry.
Protein Sci., 4, 1995
|
|
1DEC
 
 | |
1TUR
 
 | | SOLUTION STRUCTURE OF TURKEY OVOMUCOID THIRD DOMAIN AS DETERMINED FROM NUCLEAR MAGNETIC RESONANCE DATA | | Descriptor: | OVOMUCOID | | Authors: | Krezel, A.M, Darba, P, Robertson, A.D, Fejzo, J, Macura, S, Markley, J.L. | | Deposit date: | 1994-07-06 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of turkey ovomucoid third domain as determined from nuclear magnetic resonance data.
J.Mol.Biol., 242, 1994
|
|
2NA4
 
 | |
1KS6
 
 | |
1TXP
 
 | |
1X93
 
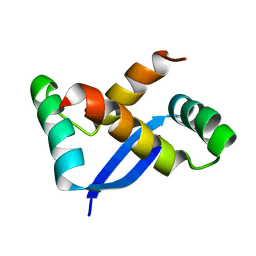 | | NMR Structure of Helicobacter pylori HP0222 | | Descriptor: | hypothetical protein HP0222 | | Authors: | Popescu, A, Karpay, A, Israel, D, Peek Jr, R.M, Krezel, A.M. | | Deposit date: | 2004-08-19 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Helicobacter pylori protein HP0222 belongs to Arc/MetJ family of transcriptional regulators.
Proteins, 59, 2005
|
|
1PGY
 
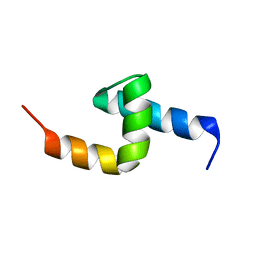 | | Solution structure of the UBA domain in Saccharomyces cerevisiae protein, Swa2p | | Descriptor: | Swa2p | | Authors: | Chim, N, Gall, W.E, Xiao, J, Harris, M.P, Graham, T.R, Krezel, A.M. | | Deposit date: | 2003-05-28 | | Release date: | 2004-03-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-binding domain in Swa2p from Saccharomyces cerevisiae.
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1TUS
 
 | |
2K1O
 
 | |
2K4J
 
 | |
2MCD
 
 | |
2MCH
 
 | |
2MAX
 
 | |
2MCK
 
 | |
1N4Y
 
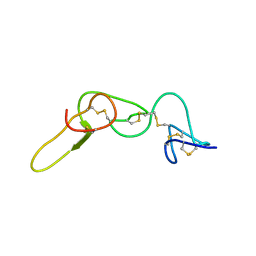 | | REFINED STRUCTURE OF KISTRIN | | Descriptor: | KISTRIN | | Authors: | Krezel, A.M, Krane, J, Dennis, M.S, Lazarus, R.A, Wagner, G. | | Deposit date: | 2002-11-02 | | Release date: | 2003-01-28 | | Last modified: | 2012-09-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist.
Science, 253, 1991
|
|
1CI6
 
 | | TRANSCRIPTION FACTOR ATF4-C/EBP BETA BZIP HETERODIMER | | Descriptor: | BETA-MERCAPTOETHANOL, FE (III) ION, TRANSCRIPTION FACTOR ATF-4, ... | | Authors: | Podust, L.M, Kim, Y. | | Deposit date: | 1999-04-07 | | Release date: | 2000-12-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA
J.Biol.Chem., 276, 2001
|
|
6WV5
 
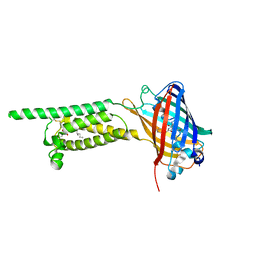 | | Human VKOR C43S mutant with vitamin K1 epoxide | | Descriptor: | (2R,3R)-2-hydroxy-3-methyl-2-[(2E,7S)-3,7,11,15-tetramethylhexadec-2-en-1-yl]-2,3-dihydronaphthalene-1,4-dione, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV6
 
 | | Human VKOR with phenindione | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phenindione, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVH
 
 | | Human VKOR with Brodifacoum | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Brodifacoum, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV3
 
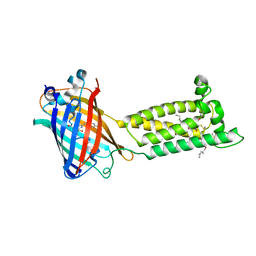 | | Human VKOR with warfarin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, S-WARFARIN, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVI
 
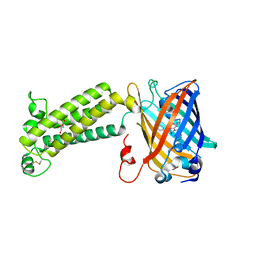 | | VKOR-like from Takifugu rubripes | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV7
 
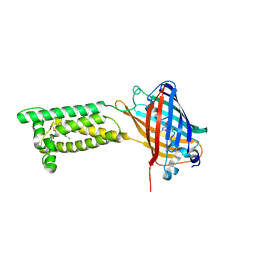 | | Human VKOR with Chlorophacinone | | Descriptor: | Chlorophacinone, Vitamin K epoxide reductase, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVB
 
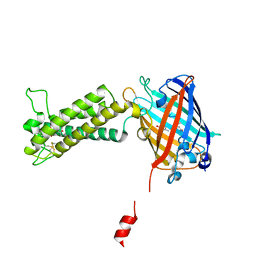 | | Takifugu rubripes VKOR-like with warfarin | | Descriptor: | S-WARFARIN, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV4
 
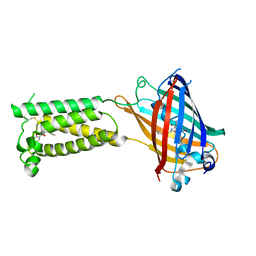 | | Human VKOR C43S with warfarin | | Descriptor: | S-WARFARIN, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.012 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
