2YOM
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Lecher, J, Hartmann, R, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
2YON
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Hartmann, R, Lecher, J, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
4FBM
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4FBL
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
3SW1
 
 | | Structure of a full-length bacterial LOV protein | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sensory box protein | | Authors: | Granzin, J, Batra-Safferling, R, Jaeger, K.-E, Drepper, T, Krauss, U. | | Deposit date: | 2011-07-13 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis for the Slow Dark Recovery of a Full-Length LOV Protein from Pseudomonas putida.
J.Mol.Biol., 417, 2012
|
|
8A33
 
 | |
4KUK
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4KUO
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
6YXB
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (space group P21) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX6
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (no morpholine) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX4
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant with morpholine | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7AB7
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) I52T Q148K variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-09-06 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7AB6
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) I52T variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-09-06 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7ABY
 
 | | Crystal structure of iLOV-Q489K mutant | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Granzin, J, Batra-Safferling, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7R4S
 
 | |
7R56
 
 | |
7R5N
 
 | |
5J3W
 
 | |
5J4E
 
 | |
6RHF
 
 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain C85A variant (CagFbFP) | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Nazarenko, V.V, Remeeva, A, Yudenko, A, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies.
Photochem. Photobiol. Sci., 18, 2019
|
|
6RHG
 
 | | Structure of Chloroflexus aggregans Cagg_3753 LOV domain | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase | | Authors: | Nazarenko, V.V, Remeeva, A, Yudenko, A, Kovalev, K, Gordeliy, V, Gushchin, I. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies.
Photochem. Photobiol. Sci., 18, 2019
|
|
5LUV
 
 | |
7YX0
 
 | | Crystal structure of the full-length short LOV protein SBW25-LOV from Pseudomonas fluorescens (light state) | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Flavin mononucleotide (semi-quinone intermediate), ... | | Authors: | Arinkin, V, Batra-Safferling, R, Granzin, J. | | Deposit date: | 2022-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved Signal Transduction Mechanisms and Dark Recovery Kinetic Tuning in the Pseudomonadaceae Short Light, Oxygen, Voltage (LOV) Protein Family.
J.Mol.Biol., 2024
|
|
8A52
 
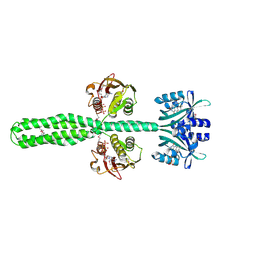 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c-axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Putative Sensory box protein,Putative Sensory box protein,Sensor protein FixL | | Authors: | Batra-Safferling, R, Arinkin, V, Granzin, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1 (asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
8A7H
 
 | | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (light state; asymmetrical variant, trigonal form with long c axis) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Flavin mononucleotide (semi-quinone intermediate), Putative Sensory box protein,Sensor protein FixL | | Authors: | Arinkin, V, Granzin, J, Batra-Safferling, R. | | Deposit date: | 2022-06-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.145 Å) | | Cite: | Crystal structure of a chimeric LOV-Histidine kinase SB2F1-I66R mutant (light state; asymmetrical variant, trigonal form with long c axis)
To Be Published
|
|
