8IFG
 
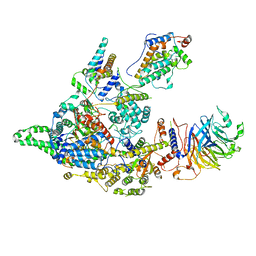 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | Authors: | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
1I6H
 
 | | RNA POLYMERASE II ELONGATION COMPLEX | | Descriptor: | 5'-D(P*AP*AP*AP*TP*GP*CP*CP*TP*GP*GP*TP*CP*T)-3', 5'-R(P*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, ... | | Authors: | Gnatt, A.L, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-03-02 | | Release date: | 2001-04-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution.
Science, 292, 2001
|
|
6XP5
 
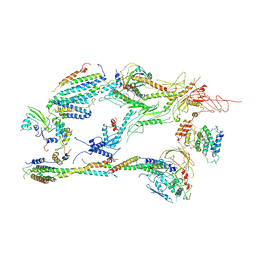 | | Head-Middle module of Mediator | | Descriptor: | HEAT, Med22, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Zhang, H.Q, Chen, D.C, Kornberg, R.D. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
3CQZ
 
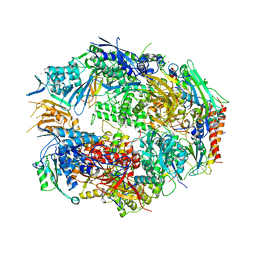 | | Crystal structure of 10 subunit RNA polymerase II in complex with the inhibitor alpha-amanitin | | Descriptor: | ALPHA-AMANITIN, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, ... | | Authors: | Kaplan, C.D, Larsson, K.-M, Kornberg, R.D. | | Deposit date: | 2008-04-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The RNA Polymerase II Trigger Loop Functions in Substrate Selection and is Directly Targeted by Alpha-Amanitin.
Mol.Cell, 30, 2008
|
|
5FMF
 
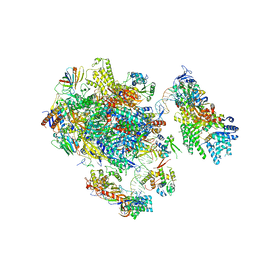 | | the P-lobe of RNA polymerase II pre-initiation complex | | Descriptor: | DNA REPAIR HELICASE RAD25, SSL2, DNA REPAIR HELICASE RAD3, ... | | Authors: | Murakami, K, Tsai, K, Kalisman, N, Bushnell, D.A, Asturias, F.J, Kornberg, R.D. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-25 | | Last modified: | 2017-07-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of an RNA Polymerase II Pre-Initiation Complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1NIK
 
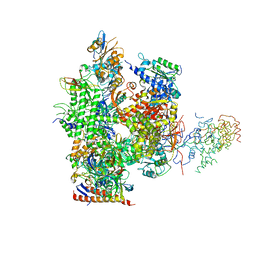 | | Wild Type RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerase I, II and III 23 kDa polypeptide, DNA-directed RNA polymerase II, ... | | Authors: | Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2002-12-24 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Complete, 12-subunit RNA Polymerase II at 4.1-A resolution: implications for the initiation of transcription.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1R9T
 
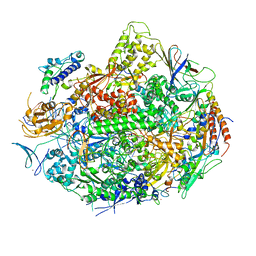 | | RNA POLYMERASE II STRAND SEPARATED ELONGATION COMPLEX, MISMATCHED NUCLEOTIDE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA nontemplate strand, DNA template strand, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1I3Q
 
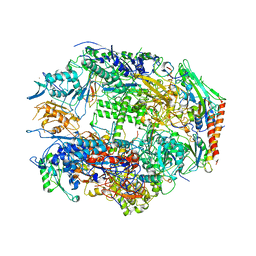 | | RNA POLYMERASE II CRYSTAL FORM I AT 3.1 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-15 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1I50
 
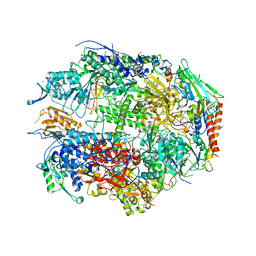 | | RNA POLYMERASE II CRYSTAL FORM II AT 2.8 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-23 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
7YFN
 
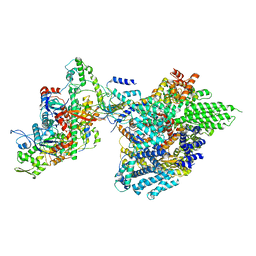 | | Core module of the NuA4 complex in S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YFP
 
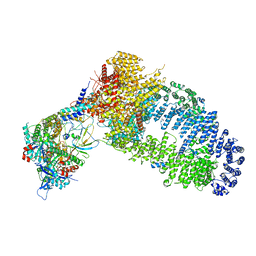 | | The NuA4 histone acetyltransferase complex from S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2E2J
 
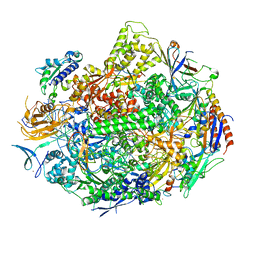 | | RNA polymerase II elongation complex in 5 mM Mg+2 with GMPCPP | | Descriptor: | 27-MER DNA template strand, 5'-D(P*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*A)-3', 5'-R(P*AP*UP*CP*GP*AP*GP*AP*GP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2E2H
 
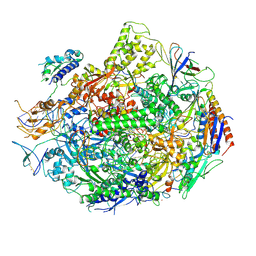 | | RNA polymerase II elongation complex at 5 mM Mg2+ with GTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2E2I
 
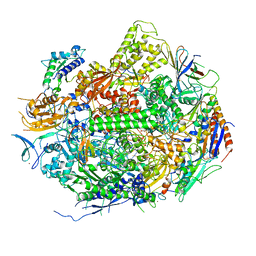 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2YU9
 
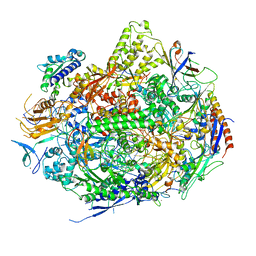 | | RNA polymerase II elongation complex in 150 mm MG+2 with UTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2007-04-06 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Transcription: Role of the Trigger Loop in Substrate Specificity and Catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
5SVA
 
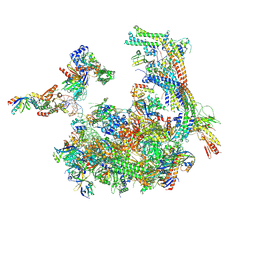 | | Mediator-RNA Polymerase II Pre-Initiation Complex | | Descriptor: | 108bp HIS4 Promoter Non-template Strand (-92/+16), 108bp HIS4 Promoter Template Strand (+16/-92), DNA repair helicase RAD25, ... | | Authors: | Robinson, P.J, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2016-08-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex.
Cell, 166, 2016
|
|
4GWP
 
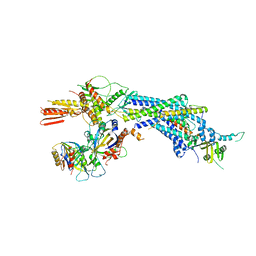 | | Structure of the Mediator Head Module from S. cerevisiae | | Descriptor: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | Authors: | Robinson, P.J.J, Bushnell, D.A, Trnka, M.J, Burlingame, A.L, Kornberg, R.D. | | Deposit date: | 2012-09-03 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GWQ
 
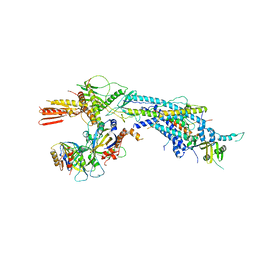 | | Structure of the Mediator Head Module from S. cerevisiae in complex with the carboxy-terminal domain (CTD) of RNA Polymerase II Rpb1 subunit | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, ... | | Authors: | Robinson, P.J.J, Bushnell, D.A, Trnka, M.J, Burlingame, A.L, Kornberg, R.D. | | Deposit date: | 2012-09-03 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1K83
 
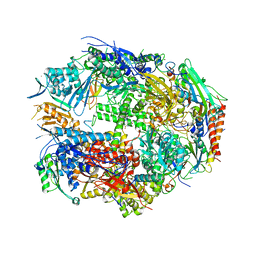 | | Crystal Structure of Yeast RNA Polymerase II Complexed with the Inhibitor Alpha Amanitin | | Descriptor: | ALPHA AMANITIN, DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, ... | | Authors: | Bushnell, D.A, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-10-22 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Transcription: Alpha-Amanitin-RNA Polymerase II Cocrystal at 2.8 A Resolution.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1R5U
 
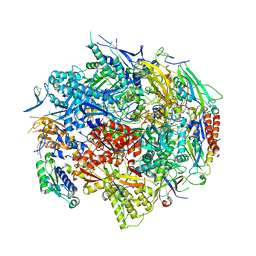 | | RNA POLYMERASE II TFIIB COMPLEX | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Bushnell, D.A, Westover, K.D, Davis, R, Kornberg, R.D. | | Deposit date: | 2003-10-13 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms.
Science, 303, 2004
|
|
1R9S
 
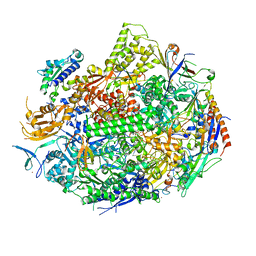 | | RNA POLYMERASE II STRAND SEPARATED ELONGATION COMPLEX, MATCHED NUCLEOTIDE | | Descriptor: | DNA strand, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SFO
 
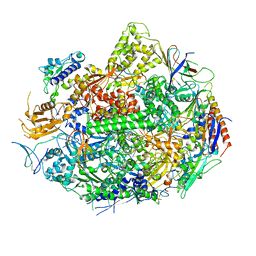 | | RNA POLYMERASE II STRAND SEPARATED ELONGATION COMPLEX | | Descriptor: | DNA STRAND, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structural Basis of Transcription: Separation of RNA from DNA by RNA Polymerase II
Science, 303, 2004
|
|
1TBP
 
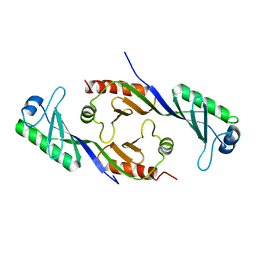 | |
1TWG
 
 | | RNA polymerase II complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1TWH
 
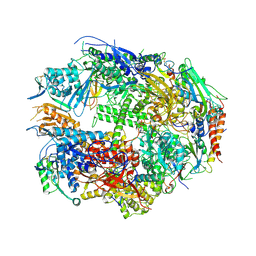 | | RNA polymerase II complexed with 2'dATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
