6DCL
 
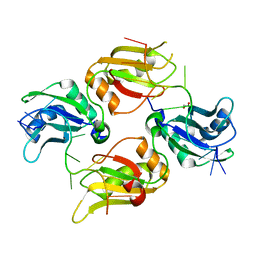 | | Crystal structure of UP1 bound to pri-miRNA-18a terminal loop | | Descriptor: | 1,2-ETHANEDIOL, Heterogeneous nuclear ribonucleoprotein A1, RNA (5'-R(*AP*GP*UP*AP*GP*AP*UP*UP*AP*GP*C)-3') | | Authors: | Kooshapur, H, Sattler, M. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1.
Nat Commun, 9, 2018
|
|
4BXU
 
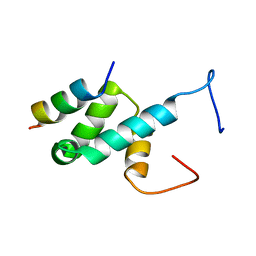 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-27 | | Last modified: | 2014-01-15 | | Method: | SOLUTION NMR | | Cite: | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
2W84
 
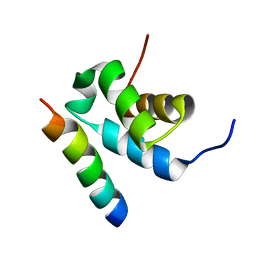 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
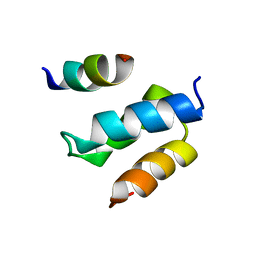 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
7KVT
 
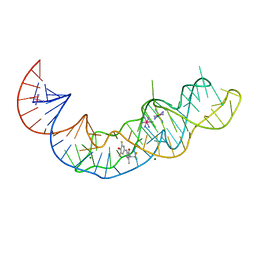 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T with iridium (III) ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVU
 
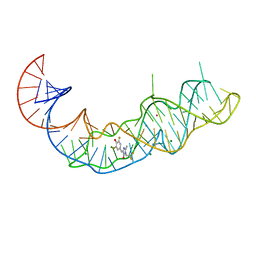 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVV
 
 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, Squash RNA aptamer bound to DFHO | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
