2G35
 
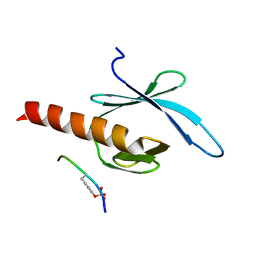 | | NMR structure of talin-PTB in complex with PIPKI | | Descriptor: | Talin-1, peptide | | Authors: | Kong, X, Wang, X, Misra, S, Qin, J. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Phosphorylation-regulated Focal Adhesion Targeting of Type Igamma Phosphatidylinositol Phosphate Kinase (PIPKIgamma) by Talin.
J.Mol.Biol., 359, 2006
|
|
6W73
 
 | |
1S4X
 
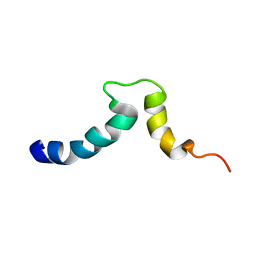 | | NMR Structure of the integrin B3 cytoplasmic domain in DPC micelles | | Descriptor: | Integrin beta-3 | | Authors: | Vinogradova, O, Vaynberg, J, Kong, X, Haas, T.A, Plow, E.F, Qin, J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Membrane-mediated structural transitions at the cytoplasmic face during integrin activation.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S4W
 
 | | NMR structure of the cytoplasmic domain of integrin AIIb in DPC micelles | | Descriptor: | Integrin alpha-IIb | | Authors: | Vinogradova, O, Vaynberg, J, Kong, X, Haas, T.A, Plow, E.F, Qin, J. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Membrane-mediated structural transitions at the cytoplasmic face during integrin activation.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3QPD
 
 | | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon | | Descriptor: | Cutinase 1 | | Authors: | Lu, A, Gosser, Y, Montclare, J.K, Liu, Z, Kong, X. | | Deposit date: | 2011-02-11 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon
To be Published
|
|
3QPA
 
 | |
3QPC
 
 | | Structure of Fusarium Solani Cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon | | Descriptor: | Cutinase | | Authors: | Lu, A, Gosser, Y, Montclare, J.K, Liu, Z, Kong, X. | | Deposit date: | 2011-02-11 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (0.978 Å) | | Cite: | Structure of Fusarium Solani Cutinase expressed in Pichia pastoris
To be Published
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | Descriptor: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
6CEZ
 
 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V2 Fab 16C2 in complex with V2 peptide ConB | | Descriptor: | HIV-1 gp120 V2 Peptide Con B, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2, Light chain of Fab fragment of rabbit anti-HIV1 gp120 V2 mAb 16C2 | | Authors: | Kong, X, Pan, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Select gp120 V2 domain specific antibodies derived from HIV and SIV infection and vaccination inhibit gp120 binding to alpha 4 beta 7.
PLoS Pathog., 14, 2018
|
|
1A7G
 
 | |
6U34
 
 | | Binary complex of native hAChE with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
4EMP
 
 | | Crystal structure of the mutant of ClpP E137A from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Zhang, J, Liu, H, Luo, C, Yang, C.-G. | | Deposit date: | 2012-04-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
J.Biol.Chem., 288, 2013
|
|
1EXZ
 
 | | STRUCTURE OF STEM CELL FACTOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, SAMARIUM (III) ION, ... | | Authors: | Zhang, Z, Zhang, R, Joachimiak, A, Schlessinger, J, Kong, X. | | Deposit date: | 2000-05-05 | | Release date: | 2000-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6O5V
 
 | | Binary complex of native hAChE with oxime reactivator RS-170B | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase.
J.Biol.Chem., 294, 2019
|
|
6O66
 
 | |
6O5R
 
 | |
6O5S
 
 | |
6U37
 
 | | Structure of VX-phosphonylated hAChE in complex with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6U3P
 
 | | Binary complex of native hAChE with oxime reactivator LG-703 | | Descriptor: | (2E,2'E)-N,N'-[1,4-diazepane-1,4-diyldi(ethane-2,1-diyl)]bis[2-(hydroxyimino)acetamide], Acetylcholinesterase, DIMETHYL SULFOXIDE | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
3GBS
 
 | | Crystal structure of Aspergillus oryzae cutinase | | Descriptor: | Cutinase 1 | | Authors: | Gosser, Y, Lu, Z, Alemu, G, Li, H, Kong, X, Liu, Z, Montclare, J. | | Deposit date: | 2009-02-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation.
J.Am.Chem.Soc., 131, 2009
|
|
4EMM
 
 | |
1N49
 
 | | Viability of a Drug-Resistant HIV-1 Protease Variant: Structural Insights for Better Anti-Viral Therapy | | Descriptor: | Protease, RITONAVIR | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Viability of a Drug-Resistant Human Immunodeficiency Virus Type 1 Protease Variant:
Structural Insights for Better Antiviral Therapy
J.VIROL., 77, 2003
|
|
4YWG
 
 | | Crystal structure of 830A in complex with V1V2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of anti-HIV-1 gp120 V1V2 antibody 830A, Light chain of anti-HIV-1 gp120 V1V2 antibody 830A, ... | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel.
J.Virol., 89, 2015
|
|
