2YQU
 
 | | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus | | Descriptor: | 2-oxoglutarate dehydrogenase E3 component, CARBONATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kondo, H, Hossain, M.T, Adachi, W, Nakai, T, Kamiya, N, Kuramitsu, K. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures and evolutionary relationship of two different lipoamide dehydrogenase(E3s) from Thermus thermophilus
To be Published
|
|
3VN3
 
 | | Fungal antifreeze protein exerts hyperactivity by constructing an inequable beta-helix | | Descriptor: | 1,2-ETHANEDIOL, Antifreeze protein | | Authors: | Kondo, H, Xiao, N, Hanada, Y, Sugimoto, H, Hoshino, T, Garnham, C.P, Davies, P.L, Tsuda, S. | | Deposit date: | 2011-12-21 | | Release date: | 2012-06-06 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Ice-binding site of snow mold fungus antifreeze protein deviates from structural regularity and high conservation
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1J05
 
 | | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment | | Descriptor: | GLYCEROL, PHOSPHATE ION, anti-CEA mAb T84.66, ... | | Authors: | Kondo, H, Nishimura, Y, Shiroishi, M, Asano, R, Noro, N, Tsumoto, K, Kumagai, I. | | Deposit date: | 2002-11-01 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment
To be Published
|
|
6A8K
 
 | |
7C3H
 
 | | Structure of L-lysine oxidase in complex with L-lysine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kondo, H, Kitagawa, M, Sugiyama, S, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
2DHT
 
 | |
1C08
 
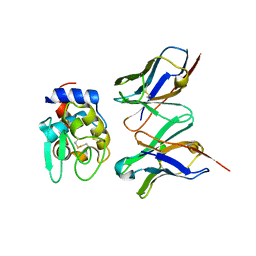 | | CRYSTAL STRUCTURE OF HYHEL-10 FV-HEN LYSOZYME COMPLEX | | Descriptor: | ANTI-HEN EGG WHITE LYSOZYME ANTIBODY (HYHEL-10), LYSOZYME | | Authors: | Shiroishi, M, Kondo, H, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1999-07-15 | | Release date: | 2000-07-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of anti-Hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex. Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain-heavy chain interfaces.
J.Biol.Chem., 274, 1999
|
|
2DCZ
 
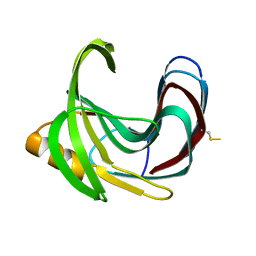 | | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase By Directed Evolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Endo-1,4-beta-xylanase A, SULFATE ION | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2DCY
 
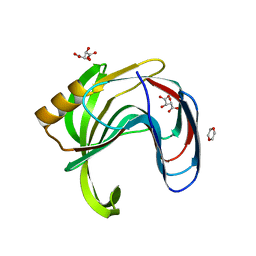 | | Crystal structure of Bacillus subtilis family-11 xylanase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D(-)-TARTARIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2E5M
 
 | |
1E32
 
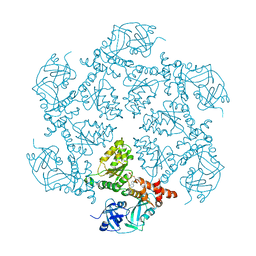 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P97 | | Authors: | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | Deposit date: | 2000-06-05 | | Release date: | 2001-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
1R7R
 
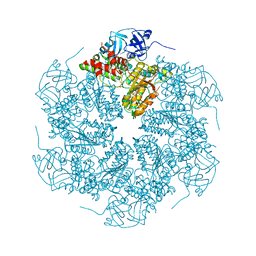 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
1S3S
 
 | | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin containing protein) (VCP) [Contains: Valosin], p47 protein | | Authors: | Dreveny, I, Kondo, H, Uchiyama, K, Shaw, A, Zhang, X, Freemont, P.S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47.
Embo J., 23, 2004
|
|
2J2M
 
 | | Crystal Structure Analysis of Catalase from Exiguobacterium oxidotolerans | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hara, I, Ichise, N, Kojima, K, Kondo, H, Ohgiya, S, Matsuyama, H, Yumoto, I. | | Deposit date: | 2006-08-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Relationship between the Size of the Bottleneck 15 a from Iron in the Main Channel and the Reactivity of Catalase Corresponding to the Molecular Size of Substrates.
Biochemistry, 46, 2007
|
|
2ZIB
 
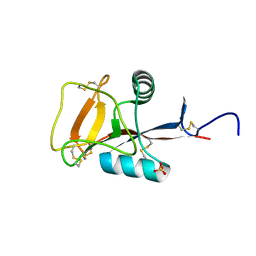 | | Crystal structure analysis of calcium-independent type II antifreeze protein | | Descriptor: | SULFATE ION, Type II antifreeze protein | | Authors: | Nishimiya, Y, Sato, R, Kondo, H, Noro, N, Sugimoto, H, Suzuki, M, Tsuda, S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure and mutational analysis of Ca2+-independent type II antifreeze protein from longsnout poacher, Brachyopsis rostratus
J.Mol.Biol., 382, 2008
|
|
3A0F
 
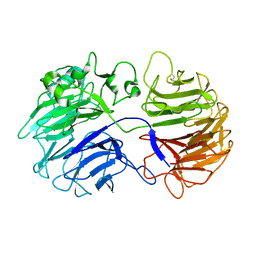 | | The crystal structure of Geotrichum sp. M128 xyloglucanase | | Descriptor: | Xyloglucanase | | Authors: | Yaoi, K, Kondo, H, Hiyoshi, A, Noro, N, Sugimoto, H, Tsuda, S, Miyazaki, K. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a xyloglucan-specific endo-beta-1,4-glucanase from Geotrichum sp. M128 xyloglucanase reveals a key amino acid residue for substrate specificity
Febs J., 276, 2009
|
|
1ULZ
 
 | | Crystal structure of the biotin carboxylase subunit of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase n-terminal domain | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Yong-Biao, J, Sueda, S, Kondo, H. | | Deposit date: | 2003-09-18 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3W3D
 
 | | Crystal structure of smooth muscle G actin DNase I complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | Authors: | Sakabe, N, Sakabe, K, Sasaki, K, Kondo, H, Shimomur, M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure and solvent network of chicken gizzard G-actin DNase 1 complex at 1.8A resolution
Acta Crystallogr.,Sect.A, 49, 1993
|
|
1J1P
 
 | | Crystal structure of HyHEL-10 Fv mutant LS91A complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
1J1O
 
 | | Crystal Structure of HyHEL-10 Fv mutant LY50F complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
1J1X
 
 | | Crystal Structure of HyHEL-10 Fv mutant LS93A complexed with hen egg white lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Yokota, A, Tsumoto, K, Shiroishi, M, Kondo, H, Kumagai, I. | | Deposit date: | 2002-12-20 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Hydrogen Bonding via Interfacial Water Molecules in Antigen-Antibody Complexation. THE HyHEL-10-HEL INTERACTION
J.BIOL.CHEM., 278, 2003
|
|
1JRU
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
2DZD
 
 | | Crystal structure of the biotin carboxylase domain of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Sueda, S, Islam, M.N, Kondo, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2EBS
 
 | | Crystal Structure Anaalysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) D465N Mutant Complexed with a Xyloglucan Heptasaccharide | | Descriptor: | Oligoxyloglucan reducing end-specific cellobiohydrolase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yaoi, K, Kondo, H, Hiyoshi, A, Noro, N, Sugimoto, H, Miyazaki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis for the Exo-mode of Action in GH74 Oligoxyloglucan Reducing End-specific Cellobiohydrolase.
J.Mol.Biol., 370, 2007
|
|
5AYU
 
 | | Crystal structure of HyHEL-10 Fv | | Descriptor: | GLYCEROL, Ig VH,anti-lysozyme, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2015-09-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of HyHEL-10 Fv
To Be Published
|
|
