2ZVY
 
 | |
5Y3Z
 
 | | Structure of the periplasmic domain of the MotB L119P mutant from Salmonella (crystal form 1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ARGININE, IMIDAZOLE, ... | | Authors: | Takao, M, Kojima, S, Sakuma, M, Homma, M, Imada, K. | | Deposit date: | 2017-07-31 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Helix Rearrangement in the Periplasmic Domain of the Flagellar Stator B Subunit Activates Peptidoglycan Binding and Ion Influx.
Structure, 26, 2018
|
|
5Y40
 
 | | Structure of the periplasmic domain of the MotB L119P mutant from Salmonella (crystal form 2) | | Descriptor: | Motility protein B | | Authors: | Takao, M, Kojima, S, Sakuma, M, Homma, M, Imada, K. | | Deposit date: | 2017-07-31 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Helix Rearrangement in the Periplasmic Domain of the Flagellar Stator B Subunit Activates Peptidoglycan Binding and Ion Influx.
Structure, 26, 2018
|
|
2ZVZ
 
 | |
2ZOV
 
 | |
2ZF8
 
 | | Crystal structure of MotY | | Descriptor: | Component of sodium-driven polar flagellar motor | | Authors: | Imada, K, Kojima, S, Namba, K, Homma, M. | | Deposit date: | 2007-12-25 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1ITP
 
 | | Solution Structure of POIA1 | | Descriptor: | proteinase A inhibitor 1 | | Authors: | Sasakawa, H, Yoshinaga, S, Kojima, S, Tamura, A. | | Deposit date: | 2002-01-23 | | Release date: | 2002-02-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of POIA1, a homologous protein to the propeptide of subtilisin: implication for protein foldability and the function as an intramolecular chaperone.
J.Mol.Biol., 317, 2002
|
|
6AHP
 
 | | Structure of the 58-167 fragment of FliL | | Descriptor: | Flagellar protein FliL | | Authors: | Takekawa, N, Isumi, M, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure ofVibrioFliL, a New Stomatin-like Protein That Assists the Bacterial Flagellar Motor Function.
MBio, 10, 2019
|
|
6AHQ
 
 | | Structure of the 40-167 fragment of FliL | | Descriptor: | Flagellar protein FliL | | Authors: | Takekawa, N, Isumi, M, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Structure ofVibrioFliL, a New Stomatin-like Protein That Assists the Bacterial Flagellar Motor Function.
MBio, 10, 2019
|
|
1IY6
 
 | | Solution structure of OMSVP3 variant, P14C/N39C | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1IY5
 
 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1V5I
 
 | | Crystal structure of serine protease inhibitor POIA1 in complex with subtilisin BPN' | | Descriptor: | CALCIUM ION, GLYCEROL, IA-1=serine proteinase inhibitor, ... | | Authors: | Lee, W.C, Kikkawa, M, Kojima, S, Miura, K, Tanokura, M. | | Deposit date: | 2003-11-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of serine protease inhibitor POIA1 in complex with subtilisin BPN'
To be Published
|
|
6IF6
 
 | | Structure of the periplasmic domain of SflA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein SflA | | Authors: | Nishikawa, S, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the periplasmic domain of SflA involved in spatial regulation of the flagellar biogenesis of Vibrio reveals a TPR/SLR-like fold.
J.Biochem., 166, 2019
|
|
7Y8X
 
 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans in complex with Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8U
 
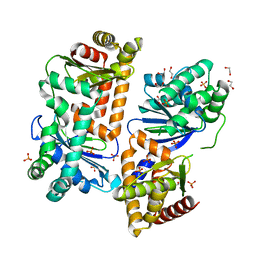 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-01-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8V
 
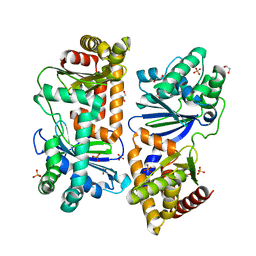 | | Crystal structure of AlbEF homolog mutant (AlbF-H54A/H58A) from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog H54A/H58A mutant, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7CIK
 
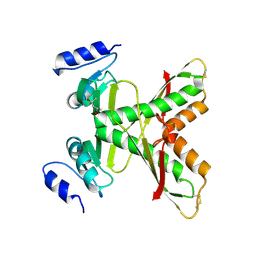 | | Structure of the 58-213 fragment of FliF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Flagellar M-ring protein | | Authors: | Takekawa, T, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-02-17 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Two Distinct Conformations in 34 FliF Subunits Generate Three Different Symmetries within the Flagellar MS-Ring.
Mbio, 12, 2021
|
|
3WPW
 
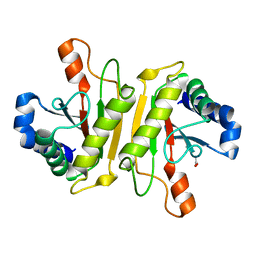 | | Structure of PomBc5, a periplasmic fragment of PomB from Vibrio | | Descriptor: | ACETATE ION, PomB | | Authors: | Takao, M, Sakuma, M, Zhu, S, Homma, M, Kojima, S, Imada, K. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational change in the periplasmic region of the flagellar stator coupled with the assembly around the rotor
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
3WPX
 
 | | Structure of PomBc4, a periplasmic fragment of PomB from Vibrio alginolyticus | | Descriptor: | PomB | | Authors: | Takao, M, Sakuma, M, Zhu, S, Homma, M, Kojima, S, Imada, K. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational change in the periplasmic region of the flagellar stator coupled with the assembly around the rotor
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
3WNZ
 
 | | Crystal structure of Bacillus subtilis YwfE, an L-amino acid ligase, with bound ADP-Mg-Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alanine-anticapsin ligase BacD, MAGNESIUM ION, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
3WO0
 
 | | Crystal structure of Bacillus subtilis YwfE, an L-amino acid ligase, with bound ADP-Mg-Ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, Alanine-anticapsin ligase BacD, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
3WO1
 
 | | Crystal structure of Trp332Ala mutant YwfE, an L-amino acid ligase, with bound ADP-Mg-Ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, Alanine-anticapsin ligase BacD, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
5SIC
 
 | |
3SIC
 
 | |
8GQY
 
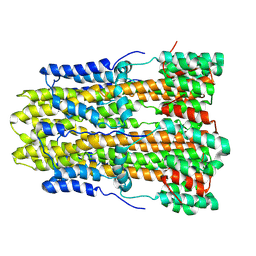 | | CryoEM structure of pentameric MotA from Aquifex aeolicus | | Descriptor: | Motility protein A | | Authors: | Nishikino, T, Takekawa, N, Kishikawa, J, Hirose, M, Onoe, S, Kato, T, Imada, K. | | Deposit date: | 2022-08-31 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of MotA, a flagellar stator protein, from hyperthermophile.
Biochem.Biophys.Res.Commun., 631, 2022
|
|
