6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | Descriptor: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6MWN
 
 | |
6U8D
 
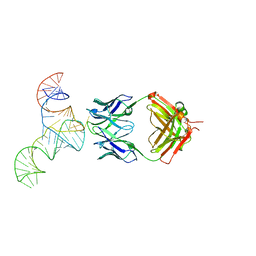 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6XJQ
 
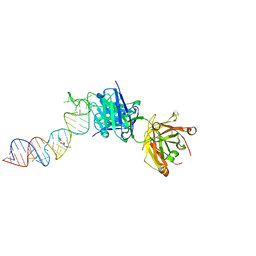 | | Crystal structure of a self-alkylating ribozyme - alkylated form with biotinylated epoxide substrate | | Descriptor: | 2-{[(4R)-4-hydroxyhexyl]oxy}ethyl 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoate, Fab HAVx Heavy Chain, Fab HAVx Light Chain, ... | | Authors: | Koirala, D, Piccirilli, J.A. | | Deposit date: | 2020-06-24 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Structural basis for substrate binding and catalysis by a self-alkylating ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
6XJY
 
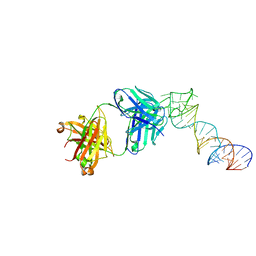 | |
6XJW
 
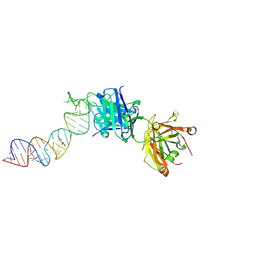 | |
6XJZ
 
 | |
6B3K
 
 | | Crystal structure of mutant Spinach RNA aptamer in complex with Fab BL3-6 | | Descriptor: | Heavy chain of Fab BL3-6, Light chain of Fab BL3-6, MAGNESIUM ION, ... | | Authors: | DasGupta, S, Koirala, D, Shelke, S.A, Piccirilli, J.A. | | Deposit date: | 2017-09-22 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography.
Nucleic Acids Res., 46, 2018
|
|
8DP3
 
 | |
8T29
 
 | |
8T2A
 
 | |
8T2B
 
 | |
8T2O
 
 | |
6B14
 
 | | Crystal structure of Spinach RNA aptamer in complex with Fab BL3-6S97N | | Descriptor: | Heavy chain of Fab BL3-6S97N, Light chain of Fab BL3-6S97N, MAGNESIUM ION, ... | | Authors: | DasGupta, S, Shelke, S.A, Piccirilli, J.A. | | Deposit date: | 2017-09-16 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography.
Nucleic Acids Res., 46, 2018
|
|
7MLX
 
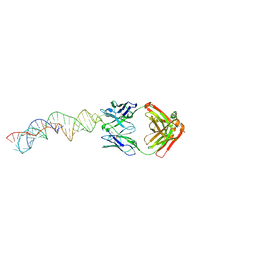 | |
6X5N
 
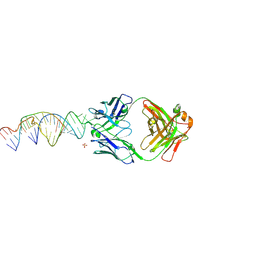 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6 | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Woldawer, A, LeGrice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
6X5M
 
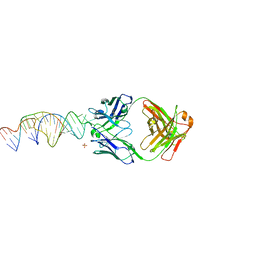 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6. | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Wlodawer, A, Le Grice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
6DB8
 
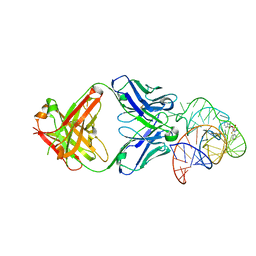 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | Descriptor: | 2-[(Z)-(3-methyl-1,3-benzoxazol-2(3H)-ylidene)methyl]-3-(3-sulfopropyl)-1,3-benzothiazol-3-ium, Fab-Heavy chain, Fab-Light chain, ... | | Authors: | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86541343 Å) | | Cite: | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
6DB9
 
 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | Descriptor: | Fab-Heavy-Chain, Fab-Light-Chain, MAGNESIUM ION, ... | | Authors: | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
7SZU
 
 | |
7U0Y
 
 | |
