2DVJ
 
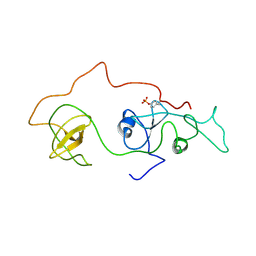 | | phosphorylated Crk-II | | Descriptor: | V-crk sarcoma virus CT10 oncogene homolog, isoform a | | Authors: | Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2006-07-31 | | Release date: | 2007-05-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYZ
 
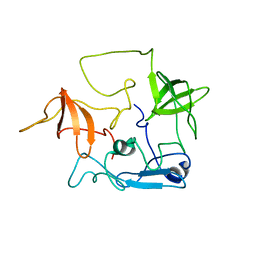 | | CT10-Regulated Kinase isoform II | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1I56
 
 | |
2EYX
 
 | |
2EYW
 
 | |
2EYV
 
 | | SH2 domain of CT10-Regulated Kinase | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYY
 
 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RSE
 
 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
3VGO
 
 | |
2LDR
 
 | |
2KE4
 
 | |
2KFK
 
 | | Solution structure of Bem1p PB1 domain complexed with Cdc24p PB1 domain | | Descriptor: | Bud emergence protein 1, Cell division control protein 24 | | Authors: | Kobashigawa, Y, Yoshinaga, S, Tandai, T, Ogura, K, Inagaki, F. | | Deposit date: | 2009-02-23 | | Release date: | 2009-10-06 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
2KBT
 
 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | Descriptor: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | Authors: | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2008-12-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
1IY3
 
 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IY4
 
 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
3VX7
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VX6
 
 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VQI
 
 | | Crystal structure of Kluyveromyces marxianus Atg5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-03-24 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
2LPU
 
 | | Solution structures of KmAtg10 | | Descriptor: | KmAtg10 | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
2LHH
 
 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2LHI
 
 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2RPV
 
 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
2KFJ
 
 | | Solution structure of the loop deletion mutant of PB1 domain of Cdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Ogura, K, Tandai, T, Yoshinaga, S, Kobashigawa, Y, Kumeta, H, Inagaki, F. | | Deposit date: | 2009-02-22 | | Release date: | 2009-10-06 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
6LCS
 
 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | Descriptor: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | Authors: | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
5B86
 
 | | Crystal structure of M-Sec | | Descriptor: | Tumor necrosis factor alpha-induced protein 2 | | Authors: | Yamashita, M, Sato, Y, Yamagata, A, Fukai, S. | | Deposit date: | 2016-06-12 | | Release date: | 2016-10-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (3.017 Å) | | Cite: | Distinct Roles for the N- and C-terminal Regions of M-Sec in Plasma Membrane Deformation during Tunneling Nanotube Formation.
Sci Rep, 6, 2016
|
|
