2IVS
 
 | | Crystal structure of non-phosphorylated RET tyrosine kinase domain | | Descriptor: | 2',3'- cyclic AMP, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Chemical Inhibition of the Ret Tyrosine Kinase Domain.
J.Biol.Chem., 281, 2006
|
|
2IVT
 
 | |
2IVV
 
 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with the inhibitor PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET PRECURSOR | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and chemical inhibition of the RET tyrosine kinase domain.
J. Biol. Chem., 281, 2006
|
|
2IVU
 
 | |
2X2L
 
 | | Crystal Structure of phosphorylated RET tyrosine kinase domain with inhibitor | | Descriptor: | (3Z)-5-AMINO-3-[(4-METHOXYPHENYL)METHYLIDENE]-1,3-DIHYDRO-2H-INDOL-2-ONE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, Kjaer, S, McDonald, N.Q. | | Deposit date: | 2010-01-13 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, structure-activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors.
Bioorg. Med. Chem., 18, 2010
|
|
2X2M
 
 | | Crystal Structure of phosphorylated RET tyrosine kinase domain with inhibitor | | Descriptor: | (3Z)-3-[(3,5-DIMETHYL-1H-PYRROL-2-YL)METHYLIDENE]-1,3-DIHYDRO-2H-INDOL-2-ONE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, Kjaer, S, McDonald, N.Q. | | Deposit date: | 2010-01-13 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis, structure-activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors.
Bioorg. Med. Chem., 18, 2010
|
|
2X2K
 
 | | Crystal Structure of phosphorylated RET tyrosine kinase domain with inhibitor | | Descriptor: | (3Z)-5-amino-3-[(3,5-dimethyl-1H-pyrrol-2-yl)methylidene]-1,3-dihydro-2H-indol-2-one, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Knowles, P.P, Murray-Rust, J, Kjaer, S, McDonald, N.Q. | | Deposit date: | 2010-01-13 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, structure-activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors.
Bioorg. Med. Chem., 18, 2010
|
|
2XQN
 
 | | Complex of the 2nd and 3rd LIM domains of TES with the EVH1 DOMAIN of MENA and the N-Terminal domain of actin-like protein Arp7A | | Descriptor: | ACTIN-LIKE PROTEIN 7A, ENABLED HOMOLOG, TESTIN, ... | | Authors: | Knowles, P.P, Briggs, D.C, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2010-09-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular recognition of the Tes LIM2-3 domains by the actin-related protein Arp7A.
J. Biol. Chem., 286, 2011
|
|
1YZB
 
 | | Solution structure of the Josephin domain of Ataxin-3 | | Descriptor: | Machado-Joseph disease protein 1 | | Authors: | Nicastro, G, Masino, L, Menon, R.P, Knowles, P.P, McDonald, N.Q, Pastore, A. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Josephin domain of ataxin-3: Structural determinants for molecular recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7AMK
 
 | | Zebrafish RET Cadherin Like Domains 1 to 4. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Purkiss, A.G, McDonald, N.Q, Goodman, K.M, Narowtek, A, Knowles, P.P. | | Deposit date: | 2020-10-09 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
3ZH8
 
 | | A novel small molecule aPKC inhibitor | | Descriptor: | (2S)-3-phenyl-N~1~-[2-(pyridin-4-yl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-yl]propane-1,2-diamine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kjaer, S, Purkiss, A.G, Kostelecky, B, Knowles, P.P, Soriano, E, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2012-12-20 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Adenosine-Binding Motif Mimicry and Cellular Effects of a Thieno[2,3-D]Pyrimidine-Based Chemical Inhibitor of Atypical Protein Kinase C Isozymes.
Biochem.J., 451, 2013
|
|
2VT8
 
 | | Structure of a conserved dimerisation domain within Fbox7 and PI31 | | Descriptor: | PROTEASOME INHIBITOR PI31 SUBUNIT | | Authors: | Kirk, R.J, Murray-Rust, J, Knowles, P.P, Laman, H, McDonald, N.Q. | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-20 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Conserved Dimerization Domain within the F-Box Protein Fbxo7 and the Pi31 Proteasome Inhibitor.
J.Biol.Chem., 283, 2008
|
|
2BHN
 
 | | XPF from Aeropyrum pernix | | Descriptor: | XPF ENDONUCLEASE | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BGW
 
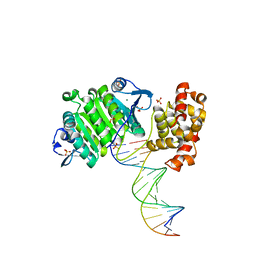 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
4UX8
 
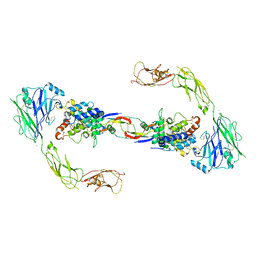 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
6FEK
 
 | |
7AB8
 
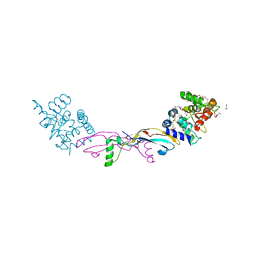 | | Crystal structure of a GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7AML
 
 | | RET/GDNF/GFRa1 extracellular complex Cryo-EM structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-10-09 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
5LI1
 
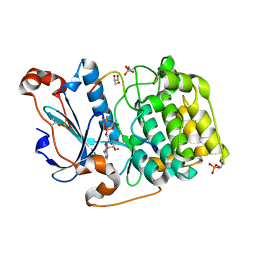 | | Structure of a Par3-inhibitory peptide bound to PKCiota core kinase domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Soriano, E.V, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
5LIH
 
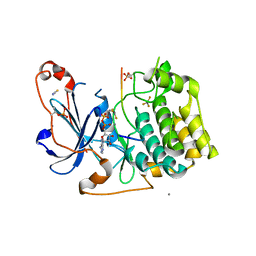 | | Structure of a peptide-substrate bound to PKCiota core kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MANGANESE (II) ION, ... | | Authors: | Soriano, E.V, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
5LI9
 
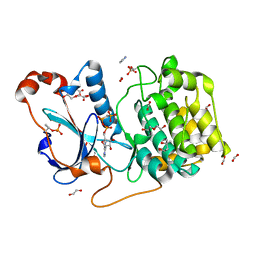 | | Structure of a nucleotide-bound form of PKCiota core kinase domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
2J16
 
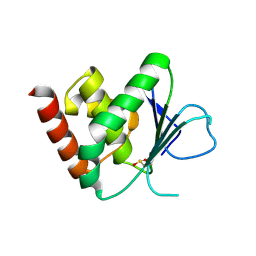 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2J17
 
 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
