7B12
 
 | | HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [2-(3-ethylphenyl)-1-[(2S)-3-phenyl-2-[(pyrazin-2-yl)formamido]propanamido]ethyl]boronic acid | | Descriptor: | ((R)-2-(3-ethylphenyl)-1-((S)-3-phenyl-2-(pyrazine-2-carboxamido)propanamido)ethyl)boronic acid, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Musil, D, Klein, M, Crosignani, S. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-01 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Optimization and Discovery of M3258, a Specific Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i).
J.Med.Chem., 64, 2021
|
|
4P1Q
 
 | | GREEN FLUORESCENT PROTEIN E222H VARIANT | | Descriptor: | Green fluorescent protein, SODIUM ION | | Authors: | Klein, M, Carius, Y, Auerbach, D, Franz, S, Jung, G, Lancaster, C.R.D. | | Deposit date: | 2014-02-27 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Replacement of Highly Conserved E222 by the Photostable Non-photoconvertible Histidine in GFP.
Chembiochem, 15, 2014
|
|
7AWE
 
 | | HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [(1R)-2-(1-benzofuran-3-yl)-1-{[(1S,2R,4R)-7-oxabicyclo[2.2.1]heptan-2-yl]formamido}ethyl]boronic acid | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Musil, D, Klein, M. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-09 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | M3258 Is a Selective Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i) Delivering Efficacy in Multiple Myeloma Models.
Mol.Cancer Ther., 20, 2021
|
|
2B90
 
 | | Crystal structure of the interleukin-4 variant T13DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8U
 
 | | Crystal structure of wildtype human Interleukin-4 | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Y
 
 | | Crystal structure of the interleukin-4 variant T13DF82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B91
 
 | | Crystal structure of the interleukin-4 variant F82DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8X
 
 | | Crystal structure of the interleukin-4 variant F82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Z
 
 | | Crystal structure of the interleukin-4 variant R85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2D48
 
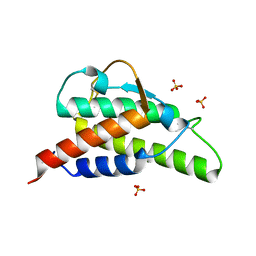 | | Crystal structure of the Interleukin-4 variant T13D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
4OD9
 
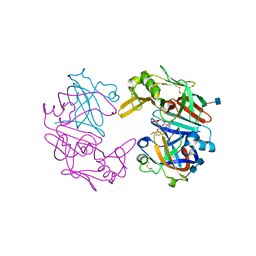 | | Structure of Cathepsin D with inhibitor N-(3,4-dimethoxybenzyl)-Nalpha-{N-[(3,4-dimethoxyphenyl)acetyl]carbamimidoyl}-D-phenylalaninamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-10 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OC6
 
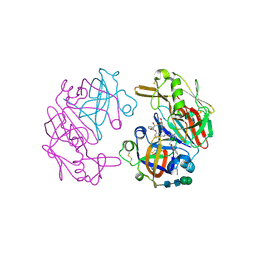 | | Structure of Cathepsin D with inhibitor 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide, Cathepsin D heavy chain, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OBZ
 
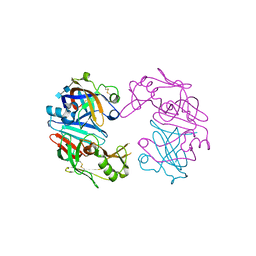 | | Structure of Cathepsin D with inhibitor 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide | | Descriptor: | 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5F9E
 
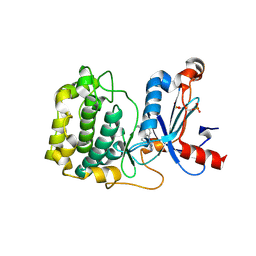 | | Structure of Protein Kinase C theta with compound 10: 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one | | Descriptor: | 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one, Protein kinase C theta type | | Authors: | Klein, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of 1,7-disubstituted-2,2-dimethyl-2,3-dihydroquinazolin-4(1H)-ones as potent and selective PKC theta inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
6XXR
 
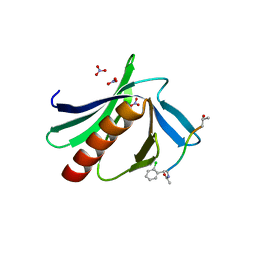 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDEA-NH2 | | Descriptor: | Ac-[2-Cl-F]-PPPPTEDEA-NH2, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7A5M
 
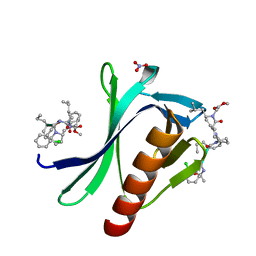 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe | | Descriptor: | Ac-[2-Cl-F]-[ProM-2]-[ProM-17]-OMe, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7AKI
 
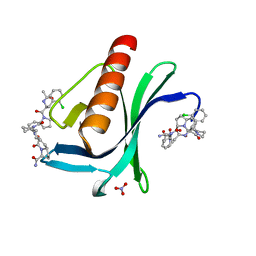 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxamide, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2020-10-01 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XVT
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDDL-NH2 | | Descriptor: | ACY-SC1-SC2-SC3-SC4-SC5-NME, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Le Cong, K, Roske, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DXL
 
 | | Linked amidobenzimidazole STING agonist | | Descriptor: | 1,1'-(butane-1,4-diyl)bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1H-benzimidazole-5-carboxamide}, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6DXG
 
 | | amidobenzimidazole (ABZI) STING agonists | | Descriptor: | 2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1-[(2R)-2-hydroxy-2-phenylethyl]-1H-benzimidazole-5-carboxamide, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
2NYT
 
 | | The APOBEC2 Crystal Structure and Functional Implications for AID | | Descriptor: | Probable C->U-editing enzyme APOBEC-2, ZINC ION | | Authors: | Prochnow, C, Bransteitter, R, Klein, M, Goodman, M, Chen, X. | | Deposit date: | 2006-11-21 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The APOBEC-2 crystal structure and functional implications for the deaminase AID.
Nature, 445, 2007
|
|
6RCF
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-15]-OH | | Descriptor: | 2-[(3~{a}~{R},6~{R},8~{a}~{S})-1-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-6-ethyl-8-oxidanylidene-3,3~{a},6,8~{a}-tetrahydro-2~{H}-pyrrolo[2,3-c]azepin-7-yl]ethanoic acid, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RCJ
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-15]-OMe | | Descriptor: | GLYCEROL, NITRATE ION, Protein enabled homolog, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RD2
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-TEDEL-NH2 | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3F9V
 
 | | Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Chen, X.J, Brewster, A.S, Wang, G.G, Yu, X, Greenleaf, W, Tjajadi, M, Klein, M. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
